Metagenomic exploration of viruses throughout the Indian Ocean - PubMed (original) (raw)
Metagenomic exploration of viruses throughout the Indian Ocean
Shannon J Williamson et al. PLoS One. 2012.
Abstract
The characterization of global marine microbial taxonomic and functional diversity is a primary goal of the Global Ocean Sampling Expedition. As part of this study, 19 water samples were collected aboard the Sorcerer II sailing vessel from the southern Indian Ocean in an effort to more thoroughly understand the lifestyle strategies of the microbial inhabitants of this ultra-oligotrophic region. No investigations of whole virioplankton assemblages have been conducted on waters collected from the Indian Ocean or across multiple size fractions thus far. Therefore, the goals of this study were to examine the effect of size fractionation on viral consortia structure and function and understand the diversity and functional potential of the Indian Ocean virome. Five samples were selected for comprehensive metagenomic exploration; and sequencing was performed on the microbes captured on 3.0-, 0.8- and 0.1 µm membrane filters as well as the viral fraction (<0.1 µm). Phylogenetic approaches were also used to identify predicted proteins of viral origin in the larger fractions of data from all Indian Ocean samples, which were included in subsequent metagenomic analyses. Taxonomic profiling of viral sequences suggested that size fractionation of marine microbial communities enriches for specific groups of viruses within the different size classes and functional characterization further substantiated this observation. Functional analyses also revealed a relative enrichment for metabolic proteins of viral origin that potentially reflect the physiological condition of host cells in the Indian Ocean including those involved in nitrogen metabolism and oxidative phosphorylation. A novel classification method, MGTAXA, was used to assess virus-host relationships in the Indian Ocean by predicting the taxonomy of putative host genera, with Prochlorococcus, Acanthochlois and members of the SAR86 cluster comprising the most abundant predictions. This is the first study to holistically explore virioplankton dynamics across multiple size classes and provides unprecedented insight into virus diversity, metabolic potential and virus-host interactions.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
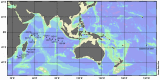
Figure 1. Map of Indian Ocean indicating where samples for metagenomic analysis were collected.
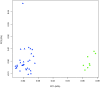
Figure 2. Principal component analysis (PCA) of the relative abundance of VF and LF viral sequences within protein clusters.
Viral libraries are represented by the green circles and larger fraction libraries are represented by the blue triangles.
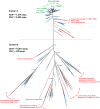
Figure 3. A phylogenetic tree built from amino acid sequences using FastTree for the gene encoding the large subunit terminase.
Selected reference sequences are colored red and green. Representative Indian Ocean VF and LF sequences are colored pink and purple respectively. GOS Phase I sequences are colored blue. Confidence values are displayed on the tree.
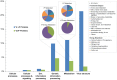
Figure 4. Functional characterization of Indian Ocean viral sequences from the viral and larger fractions of metagenomic data in the context of KEGG pathways.
The inset pie charts represent the breakdown of the Metabolism super-pathway (top) and Energy metabolism pathway (bottom). The percentages of viral sequences attributed to the Energy metabolism pathway are indicated on the Metabolism pie charts.
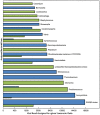
Figure 5. Bar chart demonstrating the predicted taxonomy of bacterial host genera for assembled Indian Ocean viral metagenomic data.
The area of each bar is proportional to the total number of viral reads contained in the contigs assigned to a particular host genus. Direct assignments are colored green and transitive assignments are colored blue. Direct assignments were made by assigning viral contigs to the host models built on NCBI RefSeq genomes. Transitive assignments were made by assigning viral contigs to models built on large bacterial contigs, and then assigning taxonomy to these bacterial contigs (as described in the text). Twenty top taxa in pairs of green and blue are shown according to the maximum count from either direct or transitive assignment method, respectively. If no bar is shown for one of the methods, the count was zero.
Similar articles
- The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples.
Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, Glass JI, Andrews-Pfannkoch C, Fadrosh D, Miller CS, Sutton G, Frazier M, Venter JC. Williamson SJ, et al. PLoS One. 2008 Jan 23;3(1):e1456. doi: 10.1371/journal.pone.0001456. PLoS One. 2008. PMID: 18213365 Free PMC article. - Metagenomic Analysis of the Indian Ocean Picocyanobacterial Community: Structure, Potential Function and Evolution.
Díez B, Nylander JA, Ininbergs K, Dupont CL, Allen AE, Yooseph S, Rusch DB, Bergman B. Díez B, et al. PLoS One. 2016 May 19;11(5):e0155757. doi: 10.1371/journal.pone.0155757. eCollection 2016. PLoS One. 2016. PMID: 27196065 Free PMC article. - Metagenomic Analysis Reveals Microbial Community Structure and Metabolic Potential for Nitrogen Acquisition in the Oligotrophic Surface Water of the Indian Ocean.
Wang Y, Liao S, Gai Y, Liu G, Jin T, Liu H, Gram L, Strube ML, Fan G, Sahu SK, Liu S, Gan S, Xie Z, Kong L, Zhang P, Liu X, Wang DZ. Wang Y, et al. Front Microbiol. 2021 Feb 18;12:518865. doi: 10.3389/fmicb.2021.518865. eCollection 2021. Front Microbiol. 2021. PMID: 33679623 Free PMC article. - Counts and sequences, observations that continue to change our understanding of viruses in nature.
Wommack KE, Nasko DJ, Chopyk J, Sakowski EG. Wommack KE, et al. J Microbiol. 2015 Mar;53(3):181-92. doi: 10.1007/s12275-015-5068-6. Epub 2015 Mar 3. J Microbiol. 2015. PMID: 25732739 Review. - Aquatic viral metagenomics: Lights and shadows.
Rastrojo A, Alcamí A. Rastrojo A, et al. Virus Res. 2017 Jul 15;239:87-96. doi: 10.1016/j.virusres.2016.11.021. Epub 2016 Nov 24. Virus Res. 2017. PMID: 27889617 Review.
Cited by
- Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome.
Bikel S, Valdez-Lara A, Cornejo-Granados F, Rico K, Canizales-Quinteros S, Soberón X, Del Pozo-Yauner L, Ochoa-Leyva A. Bikel S, et al. Comput Struct Biotechnol J. 2015 Jun 9;13:390-401. doi: 10.1016/j.csbj.2015.06.001. eCollection 2015. Comput Struct Biotechnol J. 2015. PMID: 26137199 Free PMC article. Review. - Over two decades of research on the marine RNA virosphere.
Liao ME, Xie Y, Shi M, Cui J. Liao ME, et al. Imeta. 2022 Oct 17;1(4):e59. doi: 10.1002/imt2.59. eCollection 2022 Dec. Imeta. 2022. PMID: 38867898 Free PMC article. Review. - Complete genome sequence of bacteriophage P8625, the first lytic phage that infects Verrucomicrobia.
Choi A, Kang I, Yang SJ, Cho JC. Choi A, et al. Stand Genomic Sci. 2015 Nov 11;10:96. doi: 10.1186/s40793-015-0091-0. eCollection 2015. Stand Genomic Sci. 2015. PMID: 26566421 Free PMC article. - Viruses in the Oceanic Basement.
Nigro OD, Jungbluth SP, Lin HT, Hsieh CC, Miranda JA, Schvarcz CR, Rappé MS, Steward GF. Nigro OD, et al. mBio. 2017 Mar 7;8(2):e02129-16. doi: 10.1128/mBio.02129-16. mBio. 2017. PMID: 28270584 Free PMC article. - Cross-Sectional Variations in Structure and Function of Coral Reef Microbiome With Local Anthropogenic Impacts on the Kenyan Coast of the Indian Ocean.
Wambua S, Gourlé H, de Villiers EP, Karlsson-Lindsjö O, Wambiji N, Macdonald A, Bongcam-Rudloff E, de Villiers S. Wambua S, et al. Front Microbiol. 2021 Jun 23;12:673128. doi: 10.3389/fmicb.2021.673128. eCollection 2021. Front Microbiol. 2021. PMID: 34248882 Free PMC article.
References
- Suttle CA (2007) Marine viruses - major players in the global ecosystem. Nature Rev Microbiol 5: 801–812. - PubMed
- Fuhrman JA (1999) Marine viruses and their biogeochemical and ecological effects. Nature 399 6736:541–548. - PubMed
- Suttle CA (2005) Viruses in the sea. Nature 437: 7057 356–61. - PubMed
- Rohwer F, Thurber RV (2009) Viruses manipulate the marine environment. Nature 459: 207–212. - PubMed
Publication types
MeSH terms
Grants and funding
This research was supported by the Office of Science (BER), U.S. Department of Energy, Cooperative Agreement No. De-FC02-02ER63453, the Gordon and Betty Moore Foundation, the National Science Foundation award 0850256 and TeraGrid allocation DEB100001 on the Texas Advanced Computing Center Ranger. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Research Materials