New concepts of endoplasmic reticulum function in the heart: programmed to conserve - PubMed (original) (raw)
Review
New concepts of endoplasmic reticulum function in the heart: programmed to conserve
Shirin Doroudgar et al. J Mol Cell Cardiol. 2013 Feb.
Abstract
Secreted and membrane proteins play critical roles in myocardial health and disease. Studies in non-myocytes have shown that the peri-nuclear ER is the site for synthesis, folding, and quality control of most secreted and membrane proteins, as well as a nexus of a signal transduction system, called the ER stress response, which informs the cell about the status of ER protein folding. Moreover, the dynamic physical and functional association of the ER with mitochondria is a key site responsible for integrating ER function and mitochondrial metabolism, but is only just beginning to be understood in the myocardium. Although a great deal is known about roles played by the sarcoplasmic reticulum (SR) in contractile calcium handling in the heart, little is known about the relative locations and functions of the peri-nuclear ER and the SR in terms of secreted and membrane protein synthesis and folding. In this review we will explore the current state of knowledge of the location of secreted and membrane protein synthesis, folding, and quality control machinery in cardiac myocytes, as well as our understanding of the functional consequences of ER stress and the unfolded protein response in the heart in terms of protein synthesis, cell growth, and metabolic regulation. This article is part of a Special Issue entitled "Focus on Cardiac Metabolism".
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
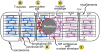
Figure 1. Sarco/endoplasmic Reticulum Network in a Cardiac Myocyte
Shown is a diagram of a cardiac myocyte depicting the relationships between the region of the SR near the t-tubule, junctional SR, the longitudinal SR (A), transverse, or t-tubules (B), the peri-nuclear ER (C) and nuclear envelope (D), and a depiction of SR that is contiguous with the nuclear envelope (E) The t-tubules are invaginations of the sarcolemma that reside over the Z-line of the sarcomeres. Also shown are the actin and myosin that comprise major portions of myofilaments, as well as the M- and Z-line regions of the sarcomeres. The nuclear envelope and peri-nuclear ER are contiguous, and constitute a location for secreted and membrane protein synthesis, as well as calcium storage and release. The hypothetical localization of secreted and membrane protein synthesis to only the nuclear envelope and peri-nuclear ER, depicted in red, and not to the SR, depicted in blue, is shown on the left of the diagram. The hypothetical localization of secreted and membrane protein synthesis to the nuclear envelope, peri-nuclear ER and the SR, depicted as a contiguous membranous system, shaded purple, is shown on the right of the diagram.
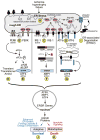
Figure 2. ER Stress Response Signaling
Shown is a diagram of the rough ER with attached ribosomes translating mRNAs that encode ER luminal proteins. Conditions that impair the folding of nascent ER proteins, which include ischemia, hypertrophy and heart failure, can result in ER stress (A). Under non-stressed conditions, the ER-luminal chaperone, glucose-regulated protein 78 kDa (GRP78) associates with the luminal domains of the 3 proximal effectors of ER stress, PERK (B), IRE-1 (C) and ATF6 (D). Upon ER stress, GRP78 relocates from the luminal domains of these proteins to misfolded proteins and either facilitates their folding, or escorts them to the degradation machinery. The disassociation of GRP78 from PERK, IRE-1 allows their oligomerization, which fosters trans-phosphorylation and activation of these effectors. In the case of ATF6, dissocation of GRP78 allows ATF6 to relocate to the Golgi, where it is cleaved by site 1 and site 2 proteases which reside in the Golgi. The resulting N-terminal fragment is liberated from the Golgi (G), translocates to the nucleus, and binds to ER stress response elements in ER stress response (ERSR) genes, and regulates their transcription. Activated PERK (C) phosphorylates the eIF2a, which fosters transient global translational repression (H) and the translation of the ATF4 mRNA from an alternate start site to generate active ATF4 using an alternate open reading frame (ORF) (E). Activated IRE-1 splices the unspliced form of XBP1 mRNA (XBP1u mRNA) to generate a splice varient form (XBP1s mRNA) which encodes the active transcription factor, XBP1 (F). Like ATF6, XBP1 and ATF6 translocate to the nucleus, and bind to various types of regulator elements in ERSRs to regulate their expression. Depending on the strength and duration of the ER stress, ERSR proteins can foster enhanced ER protein folding capacity, as well as energy conservation, which is sometimes called the adaptive ER stress response, which supports cell survival (J), or ER stress response proteins can lead to cell death, which is sometimes called the maladaptive ER stress response (K).
Similar articles
- Roles for ATF6 and the sarco/endoplasmic reticulum protein quality control system in the heart.
Glembotski CC. Glembotski CC. J Mol Cell Cardiol. 2014 Jun;71:11-5. doi: 10.1016/j.yjmcc.2013.09.018. Epub 2013 Oct 16. J Mol Cell Cardiol. 2014. PMID: 24140798 Free PMC article. Review. - Biology of endoplasmic reticulum stress in the heart.
Groenendyk J, Sreenivasaiah PK, Kim DH, Agellon LB, Michalak M. Groenendyk J, et al. Circ Res. 2010 Nov 12;107(10):1185-97. doi: 10.1161/CIRCRESAHA.110.227033. Circ Res. 2010. PMID: 21071716 Review. - Roles for the sarco-/endoplasmic reticulum in cardiac myocyte contraction, protein synthesis, and protein quality control.
Glembotski CC. Glembotski CC. Physiology (Bethesda). 2012 Dec;27(6):343-50. doi: 10.1152/physiol.00034.2012. Physiology (Bethesda). 2012. PMID: 23223628 Review. - Endoplasmic reticulum stress and diabetic cardiomyopathy.
Xu J, Zhou Q, Xu W, Cai L. Xu J, et al. Exp Diabetes Res. 2012;2012:827971. doi: 10.1155/2012/827971. Epub 2011 Nov 24. Exp Diabetes Res. 2012. PMID: 22144992 Free PMC article. Review. - Endoplasmic reticulum proteins in cardiac development and dysfunction.
Prins D, Michalak M. Prins D, et al. Can J Physiol Pharmacol. 2009 Jun;87(6):419-25. doi: 10.1139/y09-032. Can J Physiol Pharmacol. 2009. PMID: 19526035 Review.
Cited by
- Ufm1-Specific Ligase Ufl1 Regulates Endoplasmic Reticulum Homeostasis and Protects Against Heart Failure.
Li J, Yue G, Ma W, Zhang A, Zou J, Cai Y, Tang X, Wang J, Liu J, Li H, Su H. Li J, et al. Circ Heart Fail. 2018 Oct;11(10):e004917. doi: 10.1161/CIRCHEARTFAILURE.118.004917. Circ Heart Fail. 2018. PMID: 30354401 Free PMC article. - Novel insights into the cardio-protective effects of FGF21 in lean and obese rat hearts.
Patel V, Adya R, Chen J, Ramanjaneya M, Bari MF, Bhudia SK, Hillhouse EW, Tan BK, Randeva HS. Patel V, et al. PLoS One. 2014 Feb 3;9(2):e87102. doi: 10.1371/journal.pone.0087102. eCollection 2014. PLoS One. 2014. PMID: 24498293 Free PMC article. - Melatonin Protects Against Neuronal Apoptosis via Suppression of the ATF6/CHOP Pathway in a Rat Model of Intracerebral Hemorrhage.
Xu W, Lu X, Zheng J, Li T, Gao L, Lenahan C, Shao A, Zhang J, Yu J. Xu W, et al. Front Neurosci. 2018 Sep 19;12:638. doi: 10.3389/fnins.2018.00638. eCollection 2018. Front Neurosci. 2018. PMID: 30283292 Free PMC article. - Acute endoplasmic reticulum stress-induced mitochondria respiratory chain damage: The role of activated calpains.
Chen Q, Li L, Samidurai A, Thompson J, Hu Y, Willard B, Lesnefsky EJ. Chen Q, et al. FASEB J. 2024 Jan 31;38(2):e23404. doi: 10.1096/fj.202301158RR. FASEB J. 2024. PMID: 38197290 Free PMC article. - Endoplasmic reticulum stress in drug- and environmental toxicant-induced liver toxicity.
Chen S, Melchior WB Jr, Guo L. Chen S, et al. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2014;32(1):83-104. doi: 10.1080/10590501.2014.881648. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2014. PMID: 24598041 Free PMC article. Review.
References
- Porter KR, Kallman FL. Significance of cell particulates as seen by electron microscopy. Ann N Y Acad Sci. 1952;54:882–91. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 HL075573/HL/NHLBI NIH HHS/United States
- HL-075573/HL/NHLBI NIH HHS/United States
- R01 HL104535/HL/NHLBI NIH HHS/United States
- HL104535/HL/NHLBI NIH HHS/United States
- HL-085577/HL/NHLBI NIH HHS/United States
- P01 HL085577/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials