A misplaced lncRNA causes brachydactyly in humans - PubMed (original) (raw)
Clinical Trial
. 2012 Nov;122(11):3990-4002.
doi: 10.1172/JCI65508. Epub 2012 Oct 24.
Affiliations
- PMID: 23093776
- PMCID: PMC3485082
- DOI: 10.1172/JCI65508
Clinical Trial
A misplaced lncRNA causes brachydactyly in humans
Philipp G Maass et al. J Clin Invest. 2012 Nov.
Abstract
Translocations are chromosomal rearrangements that are frequently associated with a variety of disease states and developmental disorders. We identified 2 families with brachydactyly type E (BDE) resulting from different translocations affecting chromosome 12p. Both translocations caused downregulation of the parathyroid hormone-like hormone (PTHLH) gene by disrupting the cis-regulatory landscape. Using chromosome conformation capturing, we identified a regulator on chromosome 12q that interacts in cis with PTHLH over a 24.4-megabase distance and in trans with the sex-determining region Y-box 9 (SOX9) gene on chromosome 17q. The element also harbored a long noncoding RNA (lncRNA). Silencing of the lncRNA, PTHLH, or SOX9 revealed a feedback mechanism involving an expression-dependent network in humans. In the BDE patients, the human lncRNA was upregulated by the disrupted chromosomal association. Moreover, the lncRNA occupancy at the PTHLH locus was reduced. Our results document what we believe to be a novel in cis- and in trans-acting DNA and lncRNA regulatory feedback element that is reciprocally regulated by coding genes. Furthermore, our findings provide a systematic and combinatorial view of how enhancers encoding lncRNAs may affect gene expression in normal development.
Figures
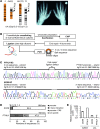
Figure 1. The translocation t(4;12)(q13.2-13.3;p11.2) and scheme of the 6C method.
(A) Translocation with der(4) and der(12). The hand roentgenogram, especially the shortened metacarpals of digits 4 and 5, are diagnostic of BDE. (B) 6C. Formaldehyde-crosslinked chromatin of C28/I2 chondrocytes was sonicated. After ChIP, the chromatin was blunt-end repaired and ligated at ultra-high dilution to favor intramolecular ligation events. Chromatin loops were PCR amplified with inverse oligos, and the amplicons were subcloned and sequenced. PTHLH and SOX9 promoters were the bait sequences. (C) Example of PTHLH 6C. Several bp were inserted due to the end repair of chromatin fragments. The interacting sequence was located at 52,431,496 bp on chromosome 12 (positions in bp; UCSC assembly hg18). In trans event in SOX9 6C. The interacting element on chromosome 12 was identified next to the bait sequence of the SOX9 promoter. (D) Restrictive classification of the 6C sequence reads. Either the promoter (bait) or the promoter interaction with any sequence was counted. (E) PTHLH and SOX9 mRNA expression in human C28/I2 and LCL.
Figure 2. 6C results and validation.
(A and B) Interaction frequencies of SOX9 (A) and PTHLH (B) 6C data. In SOX9 6C, 4 CREs were detected; 9 were found in PTHLH 6C (black symbols). re70373, re18527, and re52431 were the most abundant, and further characterized by high mammalian conservation, H3K4me1 enrichment (ENCODE), and ESPERR regulatory potential (denoted by “+”) (positions in bp; UCSC assembly hg18). re52431 interacted with SOX9 in trans. (C) New 6C libraries validated prior 6C results; control was a LCL. A direct PCR approach amplified re70373, re18527, and re52431. In the LCL, the C28/I2 CRE interactions were significantly reduced; only for re70373 were interactions in C28/I2 and LCL detected equally. (D) Distances of _cis_- and _trans_-regulatory elements identified in PTHLH 6C and SOX9 6C. re18527 (18,527,200 bp) and re52431 (52,431,500 bp) interacted with PTHLH, re70373 (70,373,400 bp) interacted in cis and re52431 in trans with SOX9. re52431 was further named CISTR-ACT, due to its cis and trans interactions. (E) Luciferase reporter assays of re70373, CISTR-ACTF (full-length regulatory sequence), and CISTR-ACTS (containing the most conserved part) elements were placed in front of the SOX9 promoter; all 3 enhanced transcription. (F) re18527, CISTR-ACTF, and CISTR-ACTS, either alone or in combination, controlled the PTHLH promoter and enhanced luciferase transcription (n = 6). ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
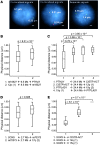
Figure 3. Colocalization-FISH.
(A) Examples of colocalized vs. separate signal pairs in C28/I2 nuclei (distances in μm; scale bars: 2 μm). (B) Accumulated colocalized signals were found in all experiments. Shown are genomic distances between probes (in Mb); Mann-Whitney test was used to determine significance. PTHLH was in physical proximity to re18527 compared with the re18527 negative control, with a 9.5-Mb equidistant upstream probe 12p(1). (C) CISTR-ACT significantly showed interaction with PTHLH compared with the 3 controls. PTPLAD1 was not expressed in C28/I2. (D) re70373 was marginally in spatial proximity to SOX9. (E) Significant in trans actions of CISTR-ACT with SOX9. In the control of SOX9 with a chromosome 15q(1) probe, only 2 signal pairs had distances less than 1 μm.
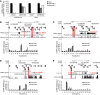
Figure 4. DA125942 transcription and dependent regulation of SOX9, DA125942, and PTHLH.
(A) CISTR-ACTS and CISTR-ACTF, with or without re18527, were cloned in a promoter and regulator lacking plasmid (pGl2-luci). In _CISTR-ACTF_–transfected C28/I2 cells, DA125942 was highly expressed. re18527 blocked the transcription; CISTR-ACTS produced no transcription. (B) SOX9 expression was knocked down relative to the scrambled siRNA control. Depleted PTHLH or DA125942 downregulated SOX9. siRNA-mediated depletion of PTHLH and SOX9 upregulated DA125942, and PTHLH expression was significantly reduced by PTHLH, DA125942, and SOX9 siRNA (n = 6). (C) Overexpression of PTHLH, SOX9, and DA125942 confirmed the expression-dependent network (n = 4). (D) STRING network. Expression array analysis revealed 22 organ morphogenesis genes (GO:0009887) showing interactions. Line thickness is indicative of physical or functional interaction confidence. Mesenchymal and prechondrogenic genes were differentially regulated. (E and F) Fibroblasts of affected BDE patients (AFF) and 3 nonaffected subjects (NON) were chondrogenically induced. DA125942 was upregulated in patients of both BDE families (n = 3). (G) RNA-ChIRP on C28/I2, nonaffected subject, and BDE patient chromatin. DA125942 was retrieved in contrast to GAPDH. (H and I) ChIRP on eluted C28/I2 DNA detected DA125942 binding at the PTHLH and SOX9 loci. Each amplicon was mapped by the UCSC custom tracks (numbered black boxes). (J and K) In t(4;12) and t(8;12), reduced lncRNA occupancy at the PTHLH locus was observed; binding at SOX9 was not affected (n = 2). ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05.
Figure 5. CISTR-ACT in rodents.
(A and B) Phylogenetic analysis of the CISTR-ACT DNA and lncRNA sequences. 121 conserved TFBSs were bioinformatically predicted. The differences of the scales indicate that the lncRNAs are evolutionarily more divergent than the conserved DNA sequences. (C) Genomic arrangement of murine and rat Pthlh, Sox9, and _CISTR-ACT_–encoded lncRNAs AW491522 and BM385392, respectively, suggested an in trans network. (D) siRNA transfections of mouse ATDC5 (10 days differentiated) chondrocytes. Depletion of the mouse lncRNA AW491522 caused Pthlh or Sox9 downregulation. Knockdown of either Pthlh or Sox9 depleted the lncRNA (n = 4). (E) siRNA assays in RCSs. An analogous expression-dependent network was observed compared with the ATDC5 results (n = 5).**P ≤ 0.01.
Figure 6. ChIRP on ATDC5 and RCS chromatin.
(A) AW491522 and BM385392 retrieval. Mouse or rat Gapdh was not precipitated. (B and C) ChIRP on eluted DNA detected AW491522 binding at the murine Pthlh (B) and Sox9 (C) loci. Each amplicon was mapped by the UCSC custom tracks (numbered black boxes). (D and E) BM385392 binding at the rat Pthlh (D) and Sox9 (E) loci. The lncRNA’s occupancy was observed at homologous regions in mouse and rat (red shading). Annotated mammalian conservation is shown; genomic positions correspond to mouse assembly mm9 and rat assembly rn4 (n = 2).
Figure 7. Transgenic embryos expressing the human and mouse CISTR-ACT_–_lacZ constructs.
(A) E10.5, E11.5, E12.5, and E13.5 embryos showed consistent lacZ expression patterns during limb bud development. fl, fore limb; hl, hind limb; d, digit; id, interdigit. Scale bars: 1 mm. (B) Results of siRNA and overexpression experiments elucidated a reciprocal expression-associated network. T-bars denote downregulation of expression; arrows denote upregulating effects. (C) Scheme of the _cis_-regulatory network between PTHLH on chromosome 12p and the CISTR-ACT locus with its lncRNA DA125942 on 12q. Translocations altered the chromosome 12 territory (right), leading to dysregulation of PTHLH and the lncRNA. Orange denotes translocated chromosomal arms.
Comment in
- Genetic "lnc"-age of noncoding RNAs to human disease.
Troy A, Sharpless NE. Troy A, et al. J Clin Invest. 2012 Nov;122(11):3837-40. doi: 10.1172/JCI66645. Epub 2012 Oct 24. J Clin Invest. 2012. PMID: 23093789 Free PMC article.
Similar articles
- A cis-regulatory site downregulates PTHLH in translocation t(8;12)(q13;p11.2) and leads to Brachydactyly Type E.
Maass PG, Wirth J, Aydin A, Rump A, Stricker S, Tinschert S, Otero M, Tsuchimochi K, Goldring MB, Luft FC, Bähring S. Maass PG, et al. Hum Mol Genet. 2010 Mar 1;19(5):848-60. doi: 10.1093/hmg/ddp553. Epub 2009 Dec 16. Hum Mol Genet. 2010. PMID: 20015959 Free PMC article. - Duplication of PTHLH causes osteochondroplasia with a combined brachydactyly type E/A1 phenotype with disturbed bone maturation and rhizomelia.
Flöttmann R, Sowinska-Seidler A, Lavie J, Chateil JF, Lacombe D, Mundlos S, Horn D, Spielmann M. Flöttmann R, et al. Eur J Hum Genet. 2016 Aug;24(8):1132-6. doi: 10.1038/ejhg.2015.266. Epub 2016 Jan 6. Eur J Hum Genet. 2016. PMID: 26733284 Free PMC article. - Report of two novel mutations in PTHLH associated with brachydactyly type E and literature review.
Thomas-Teinturier C, Pereda A, Garin I, Diez-Lopez I, Linglart A, Silve C, de Nanclares GP. Thomas-Teinturier C, et al. Am J Med Genet A. 2016 Mar;170(3):734-42. doi: 10.1002/ajmg.a.37490. Epub 2015 Dec 6. Am J Med Genet A. 2016. PMID: 26640227 Review. - Variable expressivity of the phenotype in two families with brachydactyly type E, craniofacial dysmorphism, short stature and delayed bone age caused by novel heterozygous mutations in the PTHLH gene.
Jamsheer A, Sowińska-Seidler A, Olech EM, Socha M, Kozłowski K, Pyrkosz A, Trzeciak T, Materna-Kiryluk A, Latos-Bieleńska A. Jamsheer A, et al. J Hum Genet. 2016 May;61(5):457-61. doi: 10.1038/jhg.2015.172. Epub 2016 Jan 14. J Hum Genet. 2016. PMID: 26763883 - Mutations in SOX9 cause both autosomal sex reversal and campomelic dysplasia.
Foster JW. Foster JW. Acta Paediatr Jpn. 1996 Aug;38(4):405-11. doi: 10.1111/j.1442-200x.1996.tb03515.x. Acta Paediatr Jpn. 1996. PMID: 8840554 Review.
Cited by
- Functional identification of cis-regulatory long noncoding RNAs at controlled false discovery rates.
Dhaka B, Zimmerli M, Hanhart D, Moser MB, Guillen-Ramirez H, Mishra S, Esposito R, Polidori T, Widmer M, García-Pérez R, Julio MK, Pervouchine D, Melé M, Chouvardas P, Johnson R. Dhaka B, et al. Nucleic Acids Res. 2024 Apr 12;52(6):2821-2835. doi: 10.1093/nar/gkae075. Nucleic Acids Res. 2024. PMID: 38348970 Free PMC article. - Long noncoding RNAs: cellular address codes in development and disease.
Batista PJ, Chang HY. Batista PJ, et al. Cell. 2013 Mar 14;152(6):1298-307. doi: 10.1016/j.cell.2013.02.012. Cell. 2013. PMID: 23498938 Free PMC article. Review. - Review: Long Noncoding RNAs in the Regulation of Inflammatory Pathways in Rheumatoid Arthritis and Osteoarthritis.
Pearson MJ, Jones SW. Pearson MJ, et al. Arthritis Rheumatol. 2016 Nov;68(11):2575-2583. doi: 10.1002/art.39759. Arthritis Rheumatol. 2016. PMID: 27214788 Free PMC article. Review. No abstract available. - RNA Function. RNA and dynamic nuclear organization.
Rinn J, Guttman M. Rinn J, et al. Science. 2014 Sep 12;345(6202):1240-1. doi: 10.1126/science.1252966. Science. 2014. PMID: 25214588 Free PMC article. No abstract available. - A novel heterozygous mutation in PTHLH causing autosomal dominant brachydactyly type E complicated with short stature.
Sun J, Yang N, Xu Z, Cheng H, Zhang X. Sun J, et al. Mol Genet Genomic Med. 2024 Feb;12(2):e2393. doi: 10.1002/mgg3.2393. Mol Genet Genomic Med. 2024. PMID: 38407575 Free PMC article.
References
- Bell J. The Treasury Of Human Inheritance. 1951. Brachydactyly and symphalangism. In: Penrose LS, ed. pp. 1–31. Cambridge, United Kingdom: University of Cambridge Press;
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials