Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells - PubMed (original) (raw)
Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells
Samuel Marguerat et al. Cell. 2012.
Abstract
Data on absolute molecule numbers will empower the modeling, understanding, and comparison of cellular functions and biological systems. We quantified transcriptomes and proteomes in fission yeast during cellular proliferation and quiescence. This rich resource provides the first comprehensive reference for all RNA and most protein concentrations in a eukaryote under two key physiological conditions. The integrated data set supports quantitative biology and affords unique insights into cell regulation. Although mRNAs are typically expressed in a narrow range above 1 copy/cell, most long, noncoding RNAs, except for a distinct subset, are tightly repressed below 1 copy/cell. Cell-cycle-regulated transcription tunes mRNA numbers to phase-specific requirements but can also bring about more switch-like expression. Proteins greatly exceed mRNAs in abundance and dynamic range, and concentrations are regulated to functional demands. Upon transition to quiescence, the proteome changes substantially, but, in stark contrast to mRNAs, proteins do not uniformly decrease but scale with cell volume.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
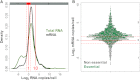
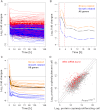
Transcriptome Quantification in Proliferating Cells (A) Abundance distribution of total RNA (green) and mRNA (black). Red vertical lines indicate 1 and 10 RNA copies/cell, and red hatched lines delimit expression zones 1 to 3. See also Figure S1 and Table S10. (B) Abundance for all detected mRNAs (each dot represents a gene). Green and gray dots correspond to essential and non essential genes, respectively. Expression zones are indicated at right.
Figure 2
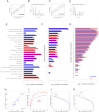
Functional Categories and Expression Zones (A) Hierarchical clustering of p values (Fisher exact test, color-coded as indicated) assessing significance of overlap between genes in functional categories (rows) and 200-gene sliding windows of mRNA abundance (columns). Vertical red lines delimit the expression zones. Functional categories with p values <0.01 in ≥1 window are shown. See also Table S11. (B) Frequency of genes for which corresponding protein is detected in 200-gene sliding window of mRNA abundance (black curve; left axis), together with p values (Fisher exact test) for significance of overlap between gene list and window (green curve; right axis). (C) As in (B) for early meiotic differentiation genes (Mata et al., 2002). (D) As in (B) for core environmental stress response genes (Chen et al., 2003). (E) As in (B) for “protein folding” genes (Gene Ontology ID: 0006457).
Figure 3
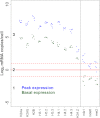
mRNA Copy Number Changes during Cell Cycle Peak (blue) and basal (green) mRNA abundance of cell-cycle-regulated genes extrapolated from average data in asynchronous cultures, with 10% of cell-cycle assumed as duration for peak expression. Data for six cell-cycle time course experiments are indicated by clustered dots (Rustici et al., 2004). Left: ten histone mRNAs peaking during S phase; right: mik1, mde6, and mei2 mRNAs peaking during M and G1 phases. See also Figure S5 and Table S12.
Figure 4
Quantitative Analysis of Long Noncoding RNAs (A) Absolute abundance of mRNAs (gray), and all (dark green), intergenic (bright green), and antisense (blue) lncRNAs. Expression zones are indicated at right. (B) Cumulative plot of copy numbers contributed by lncRNA genes ranked by decreasing abundance, with genes expressed in zones 2 and 3 at left of red line. (C) Sequence scores for lncRNAs in libraries made from total versus poly(A)+ RNA. Bright green circles, lncRNAs expressed in zone 1; dark green and orange triangles, lncRNAs expressed in zones 2 or 3; orange, lncRNAs that are ≥4-fold more abundant in total than in poly(A)+ RNA library. See also Table S13.
Figure 5
Quantitative Analysis of Proteome in Proliferating Cells (A) Abundance distribution for mRNAs (green) and proteins (red). Red vertical lines delimit expression zone 2 (0.5–2 mRNA copies/cell). See also Figures S2, S3, and Table S10. (B) Absolute abundance for all mRNAs (each dot represents a gene). Dark and light blue dots correspond to genes for which proteins were detected or not, respectively. (C) Protein versus mRNA abundance. Black curve, sliding median. (D) Protein/mRNA ratio versus protein abundance. Red dots: ribosomal proteins; black curve: sliding median. (E) Protein abundance for selected functional categories. Each dot represents a protein. Haploid Schizosaccharomyces pombe cells contain 5,110 and 10,220 annotated protein-coding genes in G1 and G2 phase, respectively (red zone), and 5,348 introns across 2,523 intron-containing genes (red and yellow zones, respectively). In proliferating cells, we measured ∼41,000 mRNA molecules (dark green line) and 1.1–2.6 × 105 copies of each rRNA (green zone). Ribosomal proteins copies/cell for paralogs were summed up. See also Figure S6.
Figure 6
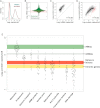
Transcriptomes and Proteomes in Proliferating versus Quiescent Cells (A) Cell volume and rRNA, mRNA, and protein copy numbers in quiescent cells as percentage of corresponding values in proliferating cells. (B) Distribution of mRNA (left) and protein (right) copies/cell during proliferation (blue) and quiescence (green), with median mRNA and protein abundance during proliferation and quiescence indicated by horizontal blue and green lines, respectively. (C) mRNA abundance in quiescent versus proliferating cells. Red and black lines delimit expression zone 2 (0.5–2 mRNA copies/cell) and 2-fold expression changes, respectively. (D) Median mRNA abundance of selected functional categories in quiescent versus proliferating cells. Red and black lines as in (C). Red and green dots indicate lowly and highly repressed categories, respectively (Table S14). (E) Protein abundance in quiescent versus proliferating cells. Black diagonal lines delimit 2-fold expression changes. (F) Median protein abundance of selected functional categories in quiescent versus proliferating cells. Black lines as in (E). Red and green dots indicate induced and repressed categories, respectively (Table S14).
Figure 7
Regulatory Dynamics during Quiescence Entry (A) Microarray time course to analyze changes in mRNA levels at 16 time points, before and 30 min to 7 days after nitrogen removal. Red profiles, mRNAs induced >1.5-fold within 3 hr after nitrogen removal; blue profiles, mRNAs repressed throughout time course. Data are normalized to 0 hr and corrected for total cellular RNA content. (B) Average expression profiles of stress- and growth-related genes, and average expression changes of all genes. (C) Absolute nCounter measurements of stress- and growth-related genes, and average profile for all 49 test genes. (D) Protein abundance in quiescent versus proliferating cells. Lower right: significance of overlap between mRNAs induced >1.5-fold within 3 hr after nitrogen removal (red dots) and proteins induced >2-fold at 24 hr after nitrogen removal.
Figure S1
Analysis of Total and Poly(dT)-Enriched Transcriptomes by Strand-Specific RNA-Seq and Calibration of RNA-Seq Data Using Absolute Measurements, Related to Figure 1 Data presented in this figure are described in detail in the Extended Experimental Procedures section. (A) Box plot of absolute reads counts in RNA-seq libraries derived from total (green) or poly(dT)-enriched (red) transcriptomes for different RNA categories. The lower and upper red lines indicate 10 and 100 sequencing reads, respectively. (B) Plot for transcript length and the correction score derived from simulated data. The red vertical lines represent, from left to right, 100, 500, and 1000 nucleotides. (C) Plot of copies of external nCounter controls used for calculation of absolute copy numbers of the 49 calibration mRNAs (nCounter run I). Grey circles represent external controls present in the nCounter mastermix. Grey triangles represent external controls added to the cellular extracts. The controls marked by a red dot were used for absolute copy number calculation. The blue dotted lines represent the most lowly and most highly expressed mRNAs for the 49 calibration genes, showing that the spikes used for copy number calculation support the whole dynamic range of the calibration set. (D) Same as (B) for nCounter run II. (E–H) Distribution of the coefficient of variations (σ/μ) of absolute copy numbers for the 49 mRNAs from the calibration set, calculated from three nCounter technical replicates split between two individual runs. (E) proliferating cells (MM1), (F) proliferating cells (MM2), (G) quiescent cells (MN1), (H) quiescent cells (MN2). (I) Plot of mRNA copies/cell for two independent biological repeats of proliferating cells (MM1 and MM2). (J) Plot of mRNA copies/cell for two independent biological repeats of quiescent cells (MN1 and MN2). (K) Natural logarithm of corrected RPK scores plotted against the natural logarithm of copies per cell for 49 mRNAs quantified by nCounter for proliferating cells. (L) Distribution of error rates determined by bootstrapping for mRNA quantities from proliferating cells. (M) Natural logarithm of corrected RPK scores plotted against the natural logarithm of copies per cell for 49 mRNAs quantified by nCounter for quiescent cells. (N) Distribution of error rates determined by bootstrapping for mRNA quantities from quiescent cells.
Figure S2
Calibration of Proteomics Data and Functional Properties of Fission Yeast Proteome, Related to Figure 5 (A) Natural logarithm of extracted precursor ion intensities plotted against the natural logarithm of copies per cell for 39 proteins quantified by heavy peptide standards for proliferating cells. (B) Distribution of error rates determined by bootstrapping for protein quantities from proliferating cells. (C) Natural logarithm of extracted precursor ion intensities plotted against the natural logarithm of copies per cell for 39 proteins quantified by heavy peptide standards for quiescent cells. (D) Distribution of error rates determined by bootstrapping for protein quantities from quiescent cells. (E) Distributions of Identified (blue bars) and all Database Protein Entries (red bars) for Clusters of orthologous groups (COG). (F) Distributions of Identified (blue bars) and all Database Protein Entries (red bars) for number of transmembrane domains per protein. (G) Distributions of Identified (blue bars) and all Database Protein Entries (red bars) for number of predicted MS-suitable peptides based on a precursor mass of 700–6000 daltons. (H) Comparison of expression levels of 17 cytokinesis proteins in asynchronous cultures as measured in this study or in a quantitative fluorescence microscopy study (Wu and Pollard, 2005). Dotted lines represent 2 and 4 fold difference. The coefficient of determination is shown in the bottom right corner. (I) Cumulative plot of the percentage of total protein count in proliferating cells as a function of the percentage expression rank of individual proteins (red curve), and of the percentage of total mRNA count as a function of the percentage expression rank of individual mRNAs (black curve). Blue and green lines mark 20 and 80% respectively. (J) Log-log plots of mRNA frequencies as a function of their expression rank. Numbers indicate the exponents of selected power-law distributions shown as black curves. The red vertical lines on the left panel delimitate the three mRNA expression zones (see main text). For more information about Pareto and Zipf laws, see (Furusawa and Kaneko, 2003; Kuznetsov et al., 2002; Newman, 2005). (K) Log-log plots of protein (right panel) frequencies as a function of their expression rank. Numbers indicate the exponents of selected power-law distributions shown as black curves. Our data thus extend Zipf's law to protein abundance.
Figure S3
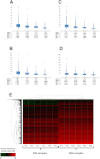
Technical and Biological Variability in Proteomics Data, Related to Figure 5 (A) Technical variability determined from replicate LC-MS/MS analyses for MM samples (proliferating). The median, mean, lower and upper endpoints of 95% confidence interval (L95 and U95) are displayed for proteins being quantified by 1, 2, 3 or more peptides. (B) Expression variability (technical and biological) between the three MM biological replicates (proliferating). (C) Technical variability determined from replicate LC-MS/MS analyses for MN samples (quiescent). The median, mean, lower and upper endpoints of 95% confidence interval (L95 and U95) are displayed for proteins being quantified by 1, 2, 3 or more peptides. (D) Expression variability (technical and biological) between the three MN biological replicates (quiescent). (E) Hierarchical clustering of absolute protein abundance in copies per cell (log10) for all 6 samples (3x MM, 3x MN) measured in duplicates each. The column dendrogram representing the clustering of the different samples is displayed.
Figure S4
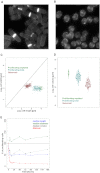
Cell Morphology during Proliferation and Quiescence, Related to Figure 6 (A) Proliferating fission yeast cells stained with calcofluor to highlight the division septa. (B) As in (A) for quiescent cells, 24h after nitrogen removal. (C) Plot of lengths and widths for 260 cells during proliferation (blue: all cells, green: septated cells), and quiescence (red). (D) Plot for distribution of cell volume for 260 cells during proliferation (blue: all cells, green: septated cells), and quiescence (red). (E) Plot for changes in cell length (blue), cell diameter (green), cell volume (gray) and total cellular RNA content (red), before and at multiple time points after nitrogen removal. Data are plotted as percentages of proliferating cells.
Figure S5
Basal and Peak Expression of Periodic Genes, Related to Figure 3 (A) Genes from M cluster, ranked according to their median basal expression levels. The horizontal red lines delimit expression zone 2 (0.5-2 mRNA copies/cell), and the three expression zones are indicated at right. (B) Same as (A) for genes from G1 cluster. (C) Same as (A) for genes from S cluster. (D) Plot of the number of genes with median basal expression switching from expression zone 1 to expression zones 2 or 3 as a function of the assumed duration of the peak phase in percent of the cell cycle for the three gene clusters (black: M phase, red: G1 phase, green: S phase). The vertical dotted line marks a ‘peak’ phase length of 10% of the cell cycle. (E) Cartoon showing three example transition patterns between ‘basal’ and ‘peak’ expression levels: An instantaneous change between the two expression states (red), expression level increases during 50% (gray) or 90% (dotted red) of the non-peak window. Ai: Amplitude of periodic variation in expression for gene i (Rustici et al., 2004). nstep: Number of intermediate states between ‘basal’ and ‘peak’ levels. (F) Impact of ramping time on the number of genes switching from zone 1 to zones 2-3 in three clusters of periodic genes as in (D), when either no ramping (red), ramping times between 10% and 80% (gray), or 90% (dotted red) of the ‘basal’ phase are incorporated in the model (nstep = 1000). The vertical dotted line marks a ‘peak’ phase length of 10% of the cell cycle.
Figure S6
Additional Analyses on Ribosomal Protein Paralogs and Transcription Factors, Related to Figure 5 (A) Expression variability of ribosomal proteins. Percent coefficient of variation in mRNA and protein expression of paralog genes from different ribosomal proteins families. Red dots: families containing repeated sequences (ambiguous mapping by RNA-seq). Blue squares: families with non-repeated sequences (un-ambiguous mapping by RNA-seq). Dots are labeled with ribosomal protein family. Black arrows: families with over 3-fold difference in protein expression between lowest and highest expressed member. Green arrows: as black arrows but for 10-fold difference. The median mRNA expression of single-copy ribosomal proteins is significantly higher than the median mRNA expression of duplicated ribosomal proteins (1.7-fold, _P_wilcox < 10−4). This patterns holds also for protein expression (1.7-fold, _P_wilcox < 0.02). As most paralogs are found in two copies, this finding suggests that each paralog might contribute to about half of the ribosomes. Fission yeast ribosomes could therefore be heterogeneous complexes with respect to paralogs. However, possible technical or classification artifacts could contribute to this observation, as RNA-seq and the proteomics approach cannot unambiguously assign expression levels to paralogs with almost identical sequences. To look in more detail into paralog expression, we calculated the percent coefficient of variation (%CV) of mRNA and protein expression for each ribosomal protein family. High %CV indicates large differences in expression between paralogs. This analysis indicates that families with low %CV, where both paralogs are likely to contribute to ribosomes, can be found in cases where sequences were sufficiently diverged to permit reliable read assignment (blue squares). Moreover, some families showed vastly divergent protein expression between paralogs (arrows), suggesting either the existence of rare specialized ribosomes or extra-ribosomal functions of the lowly expressed paralogs. (B) Comparison of TF expression levels with the occurrence of their DNA-binding motifs in the genome. Expression levels of TFs for which the DNA-binding motifs are available in PomBase were plotted against the number of their respective motifs found in the genome. Each dot represents a TF with its common name indicated in blue. It is problematic to localize true TF binding sites based on genome sequence alone, and ChIP-chip or ChIP-seq data are required to identify accurately the number of functionally relevant motifs.
Figure S7
Functional Categories and Protein Copy Numbers, Related to Figure 5 (A) Hierarchical cluster of the percentage overlap between different functional categories and sliding windows of 200 proteins of increasing abundance. Categories with at least one window containing > 15% of the proteins in a category are plotted. (B) Hierarchical cluster of the p-values of Fisher exact tests assessing the significance of the overlap between different functional categories and sliding windows of 200 proteins of increasing abundance.
Comment in
- Gene expression: dynamic omics responses.
Burgess DJ. Burgess DJ. Nat Rev Genet. 2012 Dec;13(12):828. doi: 10.1038/nrg3370. Epub 2012 Nov 14. Nat Rev Genet. 2012. PMID: 23150041 No abstract available.
Similar articles
- Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission Yeast).
Carpy A, Krug K, Graf S, Koch A, Popic S, Hauf S, Macek B. Carpy A, et al. Mol Cell Proteomics. 2014 Aug;13(8):1925-36. doi: 10.1074/mcp.M113.035824. Epub 2014 Apr 23. Mol Cell Proteomics. 2014. PMID: 24763107 Free PMC article. - Proteome analysis of Schizosaccharomyces pombe by two-dimensional gel electrophoresis and mass spectrometry.
Hwang KH, Carapito C, Böhmer S, Leize E, Van Dorsselaer A, Bernhardt R. Hwang KH, et al. Proteomics. 2006 Jul;6(14):4115-29. doi: 10.1002/pmic.200500863. Proteomics. 2006. PMID: 16791824 - Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast.
Lackner DH, Schmidt MW, Wu S, Wolf DA, Bähler J. Lackner DH, et al. Genome Biol. 2012 Apr 18;13(4):R25. doi: 10.1186/gb-2012-13-4-r25. Genome Biol. 2012. PMID: 22512868 Free PMC article. - The selective elimination of messenger RNA underlies the mitosis-meiosis switch in fission yeast.
Yamamoto M. Yamamoto M. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86(8):788-97. doi: 10.2183/pjab.86.788. Proc Jpn Acad Ser B Phys Biol Sci. 2010. PMID: 20948174 Free PMC article. Review. - Meiosis. New roles for RNA in fission yeast.
MacNeill S, Fantes P. MacNeill S, et al. Curr Biol. 1995 Jan 1;5(1):4-6. doi: 10.1016/s0960-9822(95)00002-9. Curr Biol. 1995. PMID: 7697346 Review.
Cited by
- Mrz1, a Novel Mitochondrial Outer Membrane RING Finger Protein, is Degraded Through the Ubiquitin-Proteasome Pathway in Schizosaccharomyces pombe.
Liu Z, Zhang P, Li M, A L, Yang G, Yu Y, Lu H, Shang J, Huang Y. Liu Z, et al. Curr Microbiol. 2022 Sep 10;79(10):309. doi: 10.1007/s00284-022-02998-z. Curr Microbiol. 2022. PMID: 36088506 - Transcriptome-wide association study of the plasma proteome reveals cis and trans regulatory mechanisms underlying complex traits.
Wittich H, Ardlie K, Taylor KD, Durda P, Liu Y, Mikhaylova A, Gignoux CR, Cho MH, Rich SS, Rotter JI; NHLBI TOPMed Consortium; Manichaikul A, Im HK, Wheeler HE. Wittich H, et al. Am J Hum Genet. 2024 Mar 7;111(3):445-455. doi: 10.1016/j.ajhg.2024.01.006. Epub 2024 Feb 5. Am J Hum Genet. 2024. PMID: 38320554 Free PMC article. - A stable microtubule array drives fission yeast polarity reestablishment upon quiescence exit.
Laporte D, Courtout F, Pinson B, Dompierre J, Salin B, Brocard L, Sagot I. Laporte D, et al. J Cell Biol. 2015 Jul 6;210(1):99-113. doi: 10.1083/jcb.201502025. Epub 2015 Jun 29. J Cell Biol. 2015. PMID: 26124291 Free PMC article. - Directed shotgun proteomics guided by saturated RNA-seq identifies a complete expressed prokaryotic proteome.
Omasits U, Quebatte M, Stekhoven DJ, Fortes C, Roschitzki B, Robinson MD, Dehio C, Ahrens CH. Omasits U, et al. Genome Res. 2013 Nov;23(11):1916-27. doi: 10.1101/gr.151035.112. Epub 2013 Jul 22. Genome Res. 2013. PMID: 23878158 Free PMC article. - Chromatin Dynamics During Entry to Quiescence and Compromised Functionality in Cancer Cells.
Dobbs OG, Coverley D. Dobbs OG, et al. Results Probl Cell Differ. 2022;70:279-294. doi: 10.1007/978-3-031-06573-6_9. Results Probl Cell Differ. 2022. PMID: 36348111
References
- Atkinson S.R., Marguerat S., Bähler J. Exploring long non-coding RNAs through sequencing. Semin. Cell Dev. Biol. 2012;23:200–205. - PubMed
- Beck M., Claassen M., Aebersold R. Comprehensive proteomics. Curr. Opin. Biotechnol. 2011;22:3–8. - PubMed
Supplemental References
- Bähler, J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A., III, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. - PubMed
- Elias, J.E., and Gygi, S.P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases