HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes - PubMed (original) (raw)
HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes
Jerome Jullien et al. Epigenetics Chromatin. 2012.
Abstract
Background: Nuclear reprogramming is potentially important as a route to cell replacement and drug discovery, but little is known about its mechanism. Nuclear transfer to eggs and oocytes attempts to identify the mechanism of this direct route towards reprogramming by natural components. Here we analyze how the reprogramming of nuclei transplanted to Xenopus oocytes exploits the incorporation of the histone variant H3.3.
Results: After nuclear transplantation, oocyte-derived H3.3 but not H3.2, is deposited on several regions of the genome including rDNA, major satellite repeats, and the regulatory regions of Oct4. This major H3.3 deposition occurs in absence of DNA replication, and is HIRA-and transcription-dependent. It is necessary for the shift from a somatic- to an oocyte-type of transcription after nuclear transfer.
Conclusions: This study demonstrates that the incorporation of histone H3.3 is an early and necessary step in the direct reprogramming of somatic cell nuclei by oocyte. It suggests that the incorporation of histone H3.3 is necessary during global changes in transcription that accompany changes in cell fate.
Figures
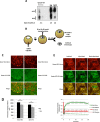
Figure 1
H3.3 but not H3.2 is accumulated in transplanted nuclei. (A) H3.2 vs. H3.3 content in oocyte GVs, eggs, and st28 embryos. Soluble pool of H3.2 and H3.3 were separated by Triton Acid Urea (TAU) gel electrophoresis and analyzed by western blot with an anti-H3 antibody. We loaded the equivalent of 20 and 10 GV, and about two eggs or st28 embryos. (B) Schematic outline of injection procedure, where mRNAs coding for tagged Xenopus histones are injected into oocyte cytoplasm 1 day before injection of donor nuclei. Oocytes containing transplanted nuclei are then incubated for 1 to 2 days and submitted to either live imaging, ChIP, or qRT-PCR analysis. (C) H3.2-cherry labelled ESC nuclei were transplanted to the germinal vesicle of H3.2-GFP or H3.3-GFP containing oocytes. Forty-eight hours after transplantation germinal vesicles were isolated and analyzed by confocal microscopy. Images are projection of Z-stack. (D) Quantification of H3.3-Cherry or H3.2-Cherry loss from transplanted nuclei. Donor ESC nuclei expressing H3.3-Cherry or H3.2-Cherry were transplanted to GVs isolated in oil and were imaged immediately (0 h) or 12 h after transplantation. The histogram shows fluorescently labeled histone signal averaged from 40 nuclei in each condition (Error bars indicate s.e.m). ★ indicate P value < 0.05, (T-TEST). (E) FRAP analysis of oocyte H3.3 and HP1alpha associated with transplanted nuclei. ESC nuclei were transplanted to H3.3-Cherry and GFP-HP1-alpha expressing oocytes. Forty-eight hours after transplantation, GVs were isolated and H3.3-cherry and GFP-HP1alpha accumulated on transplanted chromatin was subjected to FRAP analysis. Images show a clump of labeled nuclei at various time points during the FRAP procedure. The graph indicates fluorescence intensity changes over time for the tested chromatin region as well as in the surrounding GVplasm. Fluorescence intensity of H3.3 Cherry on transplanted chromatin at the beginning of the experiment is set to 1. Note that H3.3 Cherry fluorescence intensity in the GVplasm amounts to about 40% of H3.3-Cherry accumulated on chromatin, and remains constant during the course of the FRAP experiment.
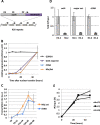
Figure 2
Kinetics of transcription and H3.3 incorporation following nuclear transplantation to Xenopus oocytes. (A) Schematic representation of the Oct4 transgene composed of a 20 copy array in the embryonic stem cells used in this study (ESC #5). DE, Distal enhancer; PE, Proximal enhancer; PP, Proximal promoter. (B) qRT-PCR analysis of transcription during a 48-h period following nuclear transplantation of 4 day-retinoic acid differentiated ESC #5 nuclei. Error bars indicate s.e.m. (n = 3). (C) Analysis of rDNA and major satellite transcription rates. Oocytes were transplanted with RA-ESCs and injected with BrUTP at various times after nuclear transfer (at 0, 12, 24, or 36 h). Oocytes were then collected 12 h after the time of BrUTP injection. In that way, the different samples collected will contain transcripts produced by the transplanted nuclei and labeled by BrUTP during a 12-h period (T12 to 24 h, T24 to 36 h, T36 to 48 h). After mRNA extraction BrUTP labeled transcripts are purified by immunoprecipitation with an anti-BrdUTP antibody and analyzed by qRT-PCR. Error bars indicate s.e.m. (n = 3). (D) ChIP analysis of histone incorporation 24 h after NT in oocytes preinjected with mRNA for HA-H3.3 and HA-H3.2, performed over the Oct4 reporter promoter, major satellite and ribosomal DNA genes, respectively. The total amount of incorporated H3.3 is set to 1. Error bars indicate standard deviation (n = 3). (E) Analysis of histone incorporation kinetics over Oct4 regulatory regions after NT. Four-day retinoic acid treated ESC #5 were transplanted in oocytes preinjected with mRNA for HA-H3.3 and HA-H3.2, collected over a 48-h time course and analyzed by ChIP with an HA antibody.
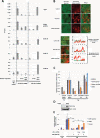
Figure 3
A HIRA- and transcription- dependent H3.3 incorporation onto transplanted chromatin is required for transcriptional reprogramming following nuclear transfer. (A) ChIP analysis of H3.2 and H3.3 incorporation onto transplanted chromatin following nuclear transfer to oocytes under conditions where transcription or HIRA is inhibited. Recipient oocytes were preinjected with either no mRNA, HA-H3.2 mRNA, or HA-H3.3 mRNA. RA-ESC #5 nuclei were then injected in the GV with or without co-injection of either alpha amanitin (RNA Polymerase II inhibition) or anti-HIRA Antibody (HIRA inhibition). Twenty-four hours after transplantation H3.2 and H3.3 incorporation onto various genomic regions was measured by ChIP analysis. Error bars indicate s.e.m. (n = 3). (B) Confocal analysis of H3.3 accumulation onto transplanted chromatin following nuclear transfer to oocytes in conditions where transcription or HIRA is inhibited. Oocytes were preinjected with H3.3-GFP mRNA. H3.2-cherry expressing ESC #5 nuclei were then transplanted to the oocyte in the presence of either alpha amanitin (RNA Polymerase II inhibition), anti-HIRA Antibody (HIRA inhibition), or control Antibody. Forty-eight hours after transplantation, H3.3-GFP loading onto chromatin was analyzed by confocal imaging of isolated GVs. Graphs show fluorescence intensity of donor H3.2-cherry (red) and oocyte H3.3-GFP (green) across a section of the image (black arrow). (C) qRT-PCR analysis of transcription 24 h following nuclear transfer of RA-ESC #5 nuclei in the presence of either alpha amanitin (RNA Polymerase II inhibition), anti-HIRA Antibody (HIRA inhibition), or control Antibody. Error bars indicate s.e.m. (n = 3). (D) Western blot analysis of xenopus HIRA expression 48 h following injection of HIRA mRNA to the oocytes. (E) qRT-PCR analysis of transcription 24 h following the transfer of RA-ESC #5 nuclei in the presence or absence of anti-HIRA antibody in GVs of oocytes pre-injected with various doses of HIRA mRNA. Error bars indicate s.e.m. (n = 3). ★ indicate P value < 0.05, (T-TEST).
Similar articles
- Histone H3 lysine 4 methylation is associated with the transcriptional reprogramming efficiency of somatic nuclei by oocytes.
Murata K, Kouzarides T, Bannister AJ, Gurdon JB. Murata K, et al. Epigenetics Chromatin. 2010 Feb 4;3(1):4. doi: 10.1186/1756-8935-3-4. Epigenetics Chromatin. 2010. PMID: 20181087 Free PMC article. - Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation.
Jullien J, Astrand C, Halley-Stott RP, Garrett N, Gurdon JB. Jullien J, et al. Proc Natl Acad Sci U S A. 2010 Mar 23;107(12):5483-8. doi: 10.1073/pnas.1000599107. Epub 2010 Mar 8. Proc Natl Acad Sci U S A. 2010. PMID: 20212135 Free PMC article. - Dramatic replacement of histone variants during genome remodeling in nuclear-transferred embryos.
Nashun B, Akiyama T, Suzuki MG, Aoki F. Nashun B, et al. Epigenetics. 2011 Dec;6(12):1489-97. doi: 10.4161/epi.6.12.18206. Epigenetics. 2011. PMID: 22139579 - H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos.
Wen D, Banaszynski LA, Rosenwaks Z, Allis CD, Rafii S. Wen D, et al. Nucleus. 2014 Sep-Oct;5(5):369-75. doi: 10.4161/nucl.36231. Nucleus. 2014. PMID: 25482190 Review. - Reprogramming of somatic cells and nuclei by Xenopus oocyte and egg extracts.
Tokmakov AA, Iwasaki T, Sato KI, Kamada S. Tokmakov AA, et al. Int J Dev Biol. 2016;60(7-8-9):289-296. doi: 10.1387/ijdb.160163at. Int J Dev Biol. 2016. PMID: 27251073 Review.
Cited by
- Histone variants and epigenetics.
Henikoff S, Smith MM. Henikoff S, et al. Cold Spring Harb Perspect Biol. 2015 Jan 5;7(1):a019364. doi: 10.1101/cshperspect.a019364. Cold Spring Harb Perspect Biol. 2015. PMID: 25561719 Free PMC article. Review. - Histone variant H3.3-mediated chromatin remodeling is essential for paternal genome activation in mouse preimplantation embryos.
Kong Q, Banaszynski LA, Geng F, Zhang X, Zhang J, Zhang H, O'Neill CL, Yan P, Liu Z, Shido K, Palermo GD, Allis CD, Rafii S, Rosenwaks Z, Wen D. Kong Q, et al. J Biol Chem. 2018 Mar 9;293(10):3829-3838. doi: 10.1074/jbc.RA117.001150. Epub 2018 Jan 22. J Biol Chem. 2018. PMID: 29358330 Free PMC article. - Histone variant H3.3 is an essential maternal factor for oocyte reprogramming.
Wen D, Banaszynski LA, Liu Y, Geng F, Noh KM, Xiang J, Elemento O, Rosenwaks Z, Allis CD, Rafii S. Wen D, et al. Proc Natl Acad Sci U S A. 2014 May 20;111(20):7325-30. doi: 10.1073/pnas.1406389111. Epub 2014 May 5. Proc Natl Acad Sci U S A. 2014. PMID: 24799717 Free PMC article. - Chromatin changes in reprogramming of mammalian somatic cells.
Xu R, Zhang S, Lei A. Xu R, et al. Rejuvenation Res. 2014 Feb;17(1):3-10. doi: 10.1089/rej.2013.1455. Rejuvenation Res. 2014. PMID: 23987213 Free PMC article. Review. - The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics.
Nie X, Wang H, Li J, Holec S, Berger F. Nie X, et al. Biol Open. 2014 Aug 1;3(9):794-802. doi: 10.1242/bio.20148680. Biol Open. 2014. PMID: 25086063 Free PMC article.
References
LinkOut - more resources
Full Text Sources