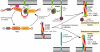The binding of Varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation - PubMed (original) (raw)
. 2012 Dec;19(12):1300-9.
doi: 10.1038/nsmb.2414. Epub 2012 Oct 28.
Affiliations
- PMID: 23104059
- PMCID: PMC3605791
- DOI: 10.1038/nsmb.2414
The binding of Varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation
Ingmar B Schäfer et al. Nat Struct Mol Biol. 2012 Dec.
Abstract
SNAREs provide energy and specificity to membrane fusion events. Fusogenic trans-SNARE complexes are assembled from glutamine-contributing SNAREs (Q-SNAREs) embedded in one membrane and an arginine-contributing SNARE (R-SNARE) embedded in the other. Regulation of membrane fusion events is crucial for intracellular trafficking. We identify the endosomal protein Varp as an R-SNARE-binding regulator of SNARE complex formation. Varp colocalizes with and binds to VAMP7, an R-SNARE that is involved in both endocytic and secretory pathways. We present the structure of the second ankyrin repeat domain of mammalian Varp in complex with the cytosolic portion of VAMP7. The VAMP7-SNARE motif is trapped between Varp and the VAMP7 longin domain, and hence Varp kinetically inhibits the ability of VAMP7 to form SNARE complexes. This inhibition will be increased when Varp can also bind to other proteins present on the same membrane as VAMP7, such as Rab32-GTP.
Figures
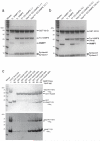
Figure 1. Mapping and confirming the binding site of Varp on VAMP7
a,c,e,f,g Coomassie–stained (Top panel) and Western blot developed with HisProbe–HRP conjugate (bottom panel) of SDS PAGE of ‘GST pull down’ assays using the baits indicated. a Full length heterologously expressed human Varp (lane ‘Varp control’) interacts with the GSTVAMP7 cytosolic domain (V7 CD) but not with GST nor the GSTVAMP7 longin domain (V7 LD) b Coomassie–stained SDS–PAGE to show effects of digesting recombinant Varp His10 with chymotrypsin (10 minutes) and trypsin (20 minutes). Proteolytic products of Varp that have an N–terminus identical to that of the full–length protein are labelled with ‘N’. The C–terminal chymotryptic fragment of Varp (residues 653–1050) marked with * has an intact C–terminal His10–tag. These products were the input for pull–down assays using GST–VAMP7CD and GST–VAMP7LD as bait. Western blotting of the SDS–PAGE from these assays developed with HisProbe–HRP conjugate showed that the fragment * can bind to the VAMP7CD. Lanes are marked Chym or Tryp for chymotryptic or tryptic digestion products, “–” for undigested. c His10-tagged residues 653–1050, 658–921 but not 718–921 of Varp can bind to the GST VAMP7CD while no Varp constructs bind to GSTVAMP7LD. d Domain organisation of human Varp (left) and mouse VAMP7 (right) as indicated by pfam [30] and in [25]. The colour scheme used is replicated in all subsequent figures. A summary of the mapping of the interaction determined using the constructs indicated is shown underneath + indicates binding and − no binding. e Varp(658–921)His6 is able to bind GSTVAMP7CD but not the GST–VAMP7 snare motif or GSTVAMP7LD. f Varp(658–921)His6 cannot bind to the GST–tagged full cytosolic domains of the mammalian longin domain containing SNAREs Sec22b and Ykt6. g Truncating GSTVAMP7 prior to residue 160 abolishes binding to Varp(658–921)His6.
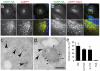
Figure 2. Varp–HA is localized to Vesicular–tubular elements of the endocytic pathway
a,b Confocal immunofluoresence of NRK cells stably expressing Varp–HA stained for the HA tag and either endogenous VAMP7 a or with anti–mRFP following transfection with mRFP–Rab7 b. Pearson R correlation coefficient for Varp–HA and VAMP7 co–localisation, 0.67 and for Varp–HA and mRFP–Rab7, 0.51. Bottom panels of a and b are zoomed in images of the boxed regions, and arrowheads indicate representative examples of structures positive for both markers. Scale bars = 20μm c,d Immunogold electron microscopy of NRK cells stably expressing Varp–HA. Dextran–Texas Red was endocytosed for 4h followed by a 20h chase to label lysosomes, and cells were then labeled with antibodies to Texas Red (5nm gold), lgp110 (10nm gold) or the HA tag (15nm gold). c Anti–HA immunoreactivity (large arrowheads show representative examples) associated with vesicular–tubular clusters (VTCs) and d dense core lysosomes co–immunolabelled with anti–Texas Red and anti–lgp110 (small arrowheads show representative examples) on ultrathin cryosections. Scale Bar = 200nm e Quantitation of the labeling density of HA associated with membranous compartments: 62% of the immunogold labeling was associated with cytosol. The relative distribution of membrane associated gold label with the plasma membrane (PM), lysosomes, endolysosomes and hybrid organelles (Endo–Lys), endosomes and vesicular–tubular clusters (E–VTC) and the trans Golgi network (TGN) is shown. Bars represent the mean of 4 independent immunolabeling experiments ± SEM.
Figure 3. Molecular details of the interaction between Varp and VAMP7
a–c Structure of the Varp–VAMP7 complex. VAMP7 (longin purple, SNARE motif in green) binds in a closed conformation to the top of the Ankyrin stack of Varp. The N–terminal Ankyrin repeat is in cyan, the four predicted Ankyrin repeats in yellow, and C–terminal Ankyrin repeat is in red. d–f Comparisons with other VAMP7 complex structures. Superposition of the VAMP7 cytosolic domain with the VAMP7–Hrb [20] and VAMP7–δ–adaptin complex [24]. The VAMP7 SNARE motif (green), the Hrb peptide (gold) and the δ–adaptin peptide (cyan) all bind to the same groove of the VAMP7 longin domain (purple). Hydrophobic residues that slot into the hydrophobic groove of VAMP7 longin domain are shown. e Structure–based alignment of the three VAMP7 longin domain binding sequences, showing that they share a pattern of hydrophobic side chains (yellow shading), but that the key residues are separated by very different sequences. f Ribbon representations of the three complexes in e superimposed on the basis of the longin domains.
Figure 4. Analysis of the Varp–VAMP7 complex interface
a Semi–transparent surface representation of the Varp–VAMP7 complex with the underlying backbone structure showing. Residues involved in the interface are shown in ball and stick representation and they and the parts of the protein surfaces they map to are highlighted in colour (VAMP7longin domain in purple, VAMP7SNARE motif in green and Varp in cyan) b The ‘flipped open’ interaction interface of the Varp–VAMP7 complex. VAMP7 and Varp have been ‘rotated’ and ‘separated’ as shown by the arrows. Side chains involved in the interaction are shown and coloured as in a with only the VAMP7 SNARE motif backbone shown for clarity. c VAMP7 SNARE motif and longin domain cytosolic domains ‘rotated’ and ‘separated’ as shown by the arrows. Residues involved in the binding to Varp are shown in darker purple (on longin domain) or green (on SNARE motif) and labelled accordingly. Residues involved in the interaction between the VAMP7 longin domain and the SNARE motif are shown in lighter purple or green and labelled in black.
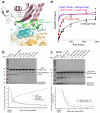
Figure 5. Mechanistic details and quantitation of the Varp–VAMP7 interaction
a The interaction interface between VAMP7 and Varp is composed of residues belonging to the first binding Ankyrin repeat of Varp and both the longin domain and the SNARE motif of VAMP7. Residues targeted by mutagenesis used to confirm the interface in solution are highlighted by a box of the same colour as the corresponding ITC trace in b and c. b and c ITC analysis of VAMP7–Varp interaction. Mutations in the Varp:VAMP7 interface abolish the binding of Varp to VAMP7. The KD measured for the wild type proteins is 2.3 ±0.6 μM and all four mutations, which do not affect folding as judged by CD (data not shown) reduce binding below detectable levels (KD>300μM).
Figure 6. The effect of Varp on VAMP7 dependent SNARE complex formation
a The trigger region of VAMP7 that initiates SNARE complex formation [41] (residues 136 LKGIMVR 142) is trapped at the centre of the Varp–VAMP7 interface. L136, I139 and V141 of the trigger region (green) are buried by interactions with the first ankyrin repeat of residues 658–921 of Varp. b and c Coomassie blue staining of SDS PAGE gels (upper panels) and quantitation by densitometry (lower panels) of i_n vitro_ SDS–stable SNARE complex reconstitution on either GSTVAMP7 (b) or on GSTVti1b SNARE motif (c). The presence of Varp residues 658–921 at the molar ratios to VAMP7 shown reduces the level of SNARE complex formed whereas the presence of the M684D,Y687S mutant form of Varp does not inhibit SNARE complex formation. d Quantitation by densitometry of Coomassie blue stained SDS PAGE gels (see Supplementary figure 8) of the time dependent effect of Varp (658–921) on SNARE complex formation on GSTVAMP7. The relative amount of SNARE complex is shown on the vertical axis (see online methods and Supplementary note). The presence of Varp (black) slows SNARE complex formation compared to the reaction in the presence of mutant Varp M684D Y687S (red). The essentially identical blue and pink curves show the equivalent time courses for the VAMP7 SNARE motif only with wt and M684D Y687S Varp present respectively.
Figure 7. Varp interacts with Rab38:GTP and VAMP7 but not VAMP8
a Coomassie blue staining of SDS PAGE gels of SNARE complex reconstitutions with the SNARE motifs of syntaxin7 and syntaxin8 on an excess of GSTVti1b for 2.5 hours using equimolar amounts (1x) of VAMP7 CD to Trx–VAMP8 alone or together. The presence of Varp(658–921)His6 at an equimolar and at a 10fold excess of the amount of VAMP7 causes significant reductions in the amount of VAMP7 incorporated into SNARE complexes as compared to the situation without Varp(658–921)His6. b Coomassie blue staining of SDS PAGE gels of SNARE complex reconstitutions as in a but using a 10–fold excess of VAMP7 CD over Trx–VAMP8. The presence of Varp(658–921)His6 again causes significant reductions in the amount of VAMP7 incorporated into SNARE complexes. ‘(Varp)’ indicates Varp(658–921)His6 that is non–specifically bound to the beads as a result of the very high amount used in the experiment. a Coomassie blue (top) and Western blotting with HisProbe–HRP conjugate of SDS–PAGE of ‘GST pull downs’ with the amounts of GTP locked (Q69L) and the GDP locked (T23N) mutant versions of GST–Rab38 indicated used as bait. VAMP7CDHis12 is only detected in pull downs on GSTRab38GTP Q69L when Varp(451–921)His6, which contains both its sets of ankyrin repeats, is also included in the reaction.
Figure8. Model of Varp function on an endosomal membrane
Through its simultaneous and direct binding to Rab32:GTP or Rab38:GTP (pink) and to VAMP7 (purple and green), Varp will trap VAMP7 in an inactive form so allowing VAMP8 (turquoise) to outcompete VAMP7 for SNARE complex formation with syntaxin7, syntaxin8 and Vti1b. Hence Varp could act to inhibit the amount of endosome:lysosome fusion and promote endosome:endosome fusion. The effect would be reversed when the GTP bound Rab was converted to its GDP bound form, which cannot bind Varp : the interaction between Varp and VAMP7 would become effectively weaker, and the resulting free, uninhibited VAMP7 could then be used in the formation of SNARE complexes that will cause endosome:lysosome fusion.
Similar articles
- VARP is recruited on to endosomes by direct interaction with retromer, where together they function in export to the cell surface.
Hesketh GG, Pérez-Dorado I, Jackson LP, Wartosch L, Schäfer IB, Gray SR, McCoy AJ, Zeldin OB, Garman EF, Harbour ME, Evans PR, Seaman MNJ, Luzio JP, Owen DJ. Hesketh GG, et al. Dev Cell. 2014 Jun 9;29(5):591-606. doi: 10.1016/j.devcel.2014.04.010. Epub 2014 May 22. Dev Cell. 2014. PMID: 24856514 Free PMC article. - Multiple Roles of VARP in Endosomal Trafficking: Rabs, Retromer Components and R-SNARE VAMP7 Meet on VARP.
Fukuda M. Fukuda M. Traffic. 2016 Jul;17(7):709-19. doi: 10.1111/tra.12406. Epub 2016 May 13. Traffic. 2016. PMID: 27103185 Review. - Structure-function analysis of VPS9-ankyrin-repeat protein (Varp) in the trafficking of tyrosinase-related protein 1 in melanocytes.
Tamura K, Ohbayashi N, Ishibashi K, Fukuda M. Tamura K, et al. J Biol Chem. 2011 Mar 4;286(9):7507-21. doi: 10.1074/jbc.M110.191205. Epub 2010 Dec 26. J Biol Chem. 2011. PMID: 21187289 Free PMC article. - The Rab21-GEF activity of Varp, but not its Rab32/38 effector function, is required for dendrite formation in melanocytes.
Ohbayashi N, Yatsu A, Tamura K, Fukuda M. Ohbayashi N, et al. Mol Biol Cell. 2012 Feb;23(4):669-78. doi: 10.1091/mbc.E11-04-0324. Epub 2011 Dec 14. Mol Biol Cell. 2012. PMID: 22171327 Free PMC article. - Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking.
Chaineau M, Danglot L, Galli T. Chaineau M, et al. FEBS Lett. 2009 Dec 3;583(23):3817-26. doi: 10.1016/j.febslet.2009.10.026. Epub 2009 Oct 20. FEBS Lett. 2009. PMID: 19837067 Review.
Cited by
- Increased activity of the vesicular soluble N-ethylmaleimide-sensitive factor attachment protein receptor TI-VAMP/VAMP7 by tyrosine phosphorylation in the Longin domain.
Burgo A, Casano AM, Kuster A, Arold ST, Wang G, Nola S, Verraes A, Dingli F, Loew D, Galli T. Burgo A, et al. J Biol Chem. 2013 Apr 26;288(17):11960-72. doi: 10.1074/jbc.M112.415075. Epub 2013 Mar 7. J Biol Chem. 2013. PMID: 23471971 Free PMC article. - Structural, Functional and Phylogenetic Analysis of Sperm Lysozyme-Like Proteins.
Kalra S, Pradeep MA, Mohanty AK, Kaushik JK. Kalra S, et al. PLoS One. 2016 Nov 10;11(11):e0166321. doi: 10.1371/journal.pone.0166321. eCollection 2016. PLoS One. 2016. PMID: 27832206 Free PMC article. - Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29.
Crawley-Snowdon H, Yang JC, Zaccai NR, Davis LJ, Wartosch L, Herman EK, Bright NA, Swarbrick JS, Collins BM, Jackson LP, Seaman MNJ, Luzio JP, Dacks JB, Neuhaus D, Owen DJ. Crawley-Snowdon H, et al. Nat Commun. 2020 Oct 6;11(1):5031. doi: 10.1038/s41467-020-18773-2. Nat Commun. 2020. PMID: 33024112 Free PMC article. - The nonphototrophic hypocotyl 3 (NPH3) domain protein NRL5 is a trafficking-associated GTPase essential for drought resistance.
Upadhyay-Tiwari N, Huang XJ, Lee YC, Singh SK, Hsu CC, Huang SS, Verslues PE. Upadhyay-Tiwari N, et al. Sci Adv. 2024 Aug 9;10(32):eado5429. doi: 10.1126/sciadv.ado5429. Epub 2024 Aug 9. Sci Adv. 2024. PMID: 39121213 Free PMC article. - Rab9A is required for delivery of cargo from recycling endosomes to melanosomes.
Mahanty S, Ravichandran K, Chitirala P, Prabha J, Jani RA, Setty SR. Mahanty S, et al. Pigment Cell Melanoma Res. 2016 Jan;29(1):43-59. doi: 10.1111/pcmr.12434. Pigment Cell Melanoma Res. 2016. PMID: 26527546 Free PMC article.
References
- Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744(3):493–517. - PubMed
- Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631–43. - PubMed
- McNew JA, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407(6801):153–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 079895/WT_/Wellcome Trust/United Kingdom
- U105178845/MRC_/Medical Research Council/United Kingdom
- MC_U105178845/MRC_/Medical Research Council/United Kingdom
- CAPMC/ CIHR/Canada
- 090909/WT_/Wellcome Trust/United Kingdom
- G0900113/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous