The σE pathway is involved in biofilm formation by Crohn's disease-associated adherent-invasive Escherichia coli - PubMed (original) (raw)
The σE pathway is involved in biofilm formation by Crohn's disease-associated adherent-invasive Escherichia coli
Benoit Chassaing et al. J Bacteriol. 2013 Jan.
Abstract
Ileal lesions of patients with Crohn's disease are colonized by adherent-invasive Escherichia coli (AIEC) bacteria that are able to adhere to and invade intestinal epithelial cells (IEC), to replicate within macrophages, and to form biofilm. Clinical observations showed that bacterial biofilms were associated with the mucosa of inflammatory bowel disease patients. In the present study, we analyzed the relationship between AIEC colonization of the gut and the formation of biofilm, focusing on the involvement of the σ(E) pathway in the AIEC-IEC interaction. We observed that σ(E) pathway inhibition in AIEC reference strain LF82 led to an impaired ability to adhere to and invade IEC but also induced a large decrease in the abilities to colonize the intestinal mucosa and form biofilm. This indicates that targeting of the σ(E) pathway could be a very potent therapeutic strategy by which to interfere with the ability of AIEC to form biofilm on the gut mucosa of Crohn's disease patients.
Figures
Fig 1
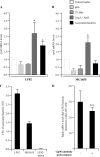
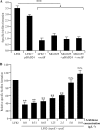
(A, B) Activation of the σE pathway in AIEC strain LF82 and nonpathogenic E. coli strain MG1655. Fold variation of rpoE mRNA levels in wild-type strains LF82 (A) and MG1655 (B) grown in medium at pH 6, in medium with 2% bile salts, or in medium with 20 g · liter−1 NaCl or adherent to I-407 epithelial cells, relative to that in wild-type strains grown in classic medium. 16S rRNA levels were measured as controls. Data are the mean ± the SEM of three separate experiments. *, P < 0.05. (C) Adhesion of AIEC strain LF82, nonpathogenic E. coli K-12 strain MG1655, and isogenic mutant LF82-Δ_fimA_ to I-407 cells. Cell-associated bacteria were quantified after a 3-h infection period. Each value is the mean number of CFU ± the SEM of at least four separate experiments. (D) Activation of the σE pathway in AIEC strain LF82 associated with I-407 cells after anti-Gp96 antibody pretreatment. Fold variation of rpoE mRNA levels in I-407 epithelial-cell-adhering bacteria of wild-type strain LF82 with or without a 30-min pretreatment of cell monolayers with anti-Gp96 antibody. N.S., not statistically significant.
Fig 2
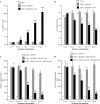
(A, B) Fold variation of rseA (A) and rpoE (B) mRNAs levels in strains LF82, LF82/pBAD24, and LF82/pBAD24-rseAB in the presence of various doses of arabinose. Results are expressed as relative expression compared to that of wild-type strain LF82 in the absence of arabinose. 16S rRNA levels were measured as controls. Data are the mean ± the SEM of three separate experiments. (C, D) Activation of the rpoE (C) and rpoH (D) promoters in strains LF82, LF82/pBAD24, and LF82/pBAD24-rseAB in the presence of various doses of arabinose. Shown is the β-galactosidase activity per OD620 unit resulting from the expression of lacZ driven by the DNA sequence upstream of the rpoE or rpoH gene. Data are the mean ± the SEM of four separate experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Fig 3
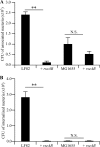
(A, B) Abilities of strains LF82, LF82/pBAD24-rseAB, MG1655, and MG1655/pBAD24-rseAB to adhere to (A) and invade (B) I-407 IEC. Each value is the mean ± the SEM of at least four separate experiments. **, P < 0.01; N.S., not statistically significant.
Fig 4
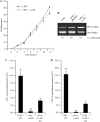
(A) Motility assay of wild-type strain LF82 and strain LF82/pBAD24-rseAB on 0.3% agar at 37°C. (B) Regulation of type 1 pili in strain LF82/pBAD24-rseAB. PCR analysis was used to determine the invertible element orientation of the fim operon in strains LF82, LF82/pBAD24, and LF82/pBAD24-rseAB. A 450-bp product revealed the ON orientation of the invertible element, and a 750-bp product revealed its OFF orientation. (C, D) The abilities of strains LF82 + pHSG575-fim, LF82/pBAD24-rseAB, and LF82/pBAD24-rseAB/pHSG575-fim to adhere to (C) and invade (D) I-407 IEC. Centrifugation was performed to force contact between bacteria and I-407 IEC. Each value is the mean ± the SEM of at least four separate experiments. *, P < 0.05; **, P < 0.01.
Fig 5
(A) SBF indexes of AIEC strain LF82 and nonpathogenic E. coli strain MG1655 with or without RseAB overexpression. Data are the mean ± the SEM of three separate experiments. (B) SBF index of isogenic mutant LF82-Δ_rpoE_ transcomplemented with pBAD30-rpoE and grown in the presence of 0.00, 0.31, 0.63, 1.25, 2.50, 5.00, or 10.00 g · liter−1 arabinose. Data are the mean ± the SEM of three separate experiments. **, P < 0.01; ***, P < 0.001; N.S., not statistically significant.
Fig 6
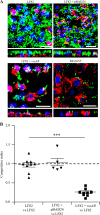
(A) Confocal analysis of LF82, LF82/pBAD24, LF82/pBAD24-rseAB, and MG1655 biofilm formation at the surface of a PFA-fixed monolayer of I-407 IEC. Bacteria expressing GFP were used, actin is stained red with phalloidin-TRITC, and nuclei are stained blue with Hoechst. Representative z sections were visualized under each confocal slice. Bars, 50 μm. (B) CI of strain LF82/pBAD24-rseAB compared to that of wild-type strain LF82. Intestinal ileal loops were inoculated with mixed inoculums comprising equivalent numbers of the wild-type and LF82 pBAD24-rseAB strains, and their presence was compared by CI analysis, which provides a sensitive measurement of the relative degree of attenuation. **, P < 0.01; ***, P < 0.001.
Fig 7
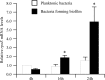
Activation of the σE pathway in AIEC strain LF82 during the biofilm formation process. Shown is the _n_-fold variation of rpoE mRNA levels in wild-type strain LF82 during biofilm formation (4, 16, and 24 h) relative to that of the wild-type strain grown for 4 h in classic medium. 16S rRNA levels were measured as controls. Data are the mean ± the SEM of three separate experiments. *, P < 0.05; **, P < 0.01.
Similar articles
- Analysis of the σE regulon in Crohn's disease-associated Escherichia coli revealed involvement of the waaWVL operon in biofilm formation.
Chassaing B, Garénaux E, Carriere J, Rolhion N, Guérardel Y, Barnich N, Bonnet R, Darfeuille-Michaud A. Chassaing B, et al. J Bacteriol. 2015 Apr;197(8):1451-65. doi: 10.1128/JB.02499-14. Epub 2015 Feb 9. J Bacteriol. 2015. PMID: 25666140 Free PMC article. - Development of Heptylmannoside-Based Glycoconjugate Antiadhesive Compounds against Adherent-Invasive Escherichia coli Bacteria Associated with Crohn's Disease.
Sivignon A, Yan X, Alvarez Dorta D, Bonnet R, Bouckaert J, Fleury E, Bernard J, Gouin SG, Darfeuille-Michaud A, Barnich N. Sivignon A, et al. mBio. 2015 Nov 17;6(6):e01298-15. doi: 10.1128/mBio.01298-15. mBio. 2015. PMID: 26578673 Free PMC article. - The oxidoreductase DsbA plays a key role in the ability of the Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 to resist macrophage killing.
Bringer MA, Rolhion N, Glasser AL, Darfeuille-Michaud A. Bringer MA, et al. J Bacteriol. 2007 Jul;189(13):4860-71. doi: 10.1128/JB.00233-07. Epub 2007 Apr 20. J Bacteriol. 2007. PMID: 17449627 Free PMC article. - Adherent-Invasive E. coli: Update on the Lifestyle of a Troublemaker in Crohn's Disease.
Chervy M, Barnich N, Denizot J. Chervy M, et al. Int J Mol Sci. 2020 May 25;21(10):3734. doi: 10.3390/ijms21103734. Int J Mol Sci. 2020. PMID: 32466328 Free PMC article. Review. - Adherent-invasive Escherichia coli in inflammatory bowel disease.
Rolhion N, Darfeuille-Michaud A. Rolhion N, et al. Inflamm Bowel Dis. 2007 Oct;13(10):1277-83. doi: 10.1002/ibd.20176. Inflamm Bowel Dis. 2007. PMID: 17476674 Review.
Cited by
- Siderophore Immunization Restricted Colonization of Adherent-Invasive Escherichia coli and Ameliorated Experimental Colitis.
Gerner RR, Hossain S, Sargun A, Siada K, Norton GJ, Zheng T, Neumann W, Nuccio SP, Nolan EM, Raffatellu M. Gerner RR, et al. mBio. 2022 Oct 26;13(5):e0218422. doi: 10.1128/mbio.02184-22. Epub 2022 Sep 12. mBio. 2022. PMID: 36094114 Free PMC article. - Professor Arlette Darfeuille-Michaud: the discovery of adherent-invasive Escherichia coli.
Yang Y, Jobin C. Yang Y, et al. J Crohns Colitis. 2015 May;9(5):373-5. doi: 10.1093/ecco-jcc/jjv044. Epub 2015 Mar 10. J Crohns Colitis. 2015. PMID: 25759068 Free PMC article. No abstract available. - Inactivation of the Pyrimidine Biosynthesis pyrD Gene Negatively Affects Biofilm Formation and Virulence Determinants in the Crohn's Disease-Associated Adherent Invasive Escherichia coli LF82 Strain.
Rossi E, Leccese G, Baldelli V, Bibi A, Scalone E, Camilloni C, Paroni M, Landini P. Rossi E, et al. Microorganisms. 2022 Feb 28;10(3):537. doi: 10.3390/microorganisms10030537. Microorganisms. 2022. PMID: 35336113 Free PMC article. - Analysis of the σE regulon in Crohn's disease-associated Escherichia coli revealed involvement of the waaWVL operon in biofilm formation.
Chassaing B, Garénaux E, Carriere J, Rolhion N, Guérardel Y, Barnich N, Bonnet R, Darfeuille-Michaud A. Chassaing B, et al. J Bacteriol. 2015 Apr;197(8):1451-65. doi: 10.1128/JB.02499-14. Epub 2015 Feb 9. J Bacteriol. 2015. PMID: 25666140 Free PMC article. - Surface-Associated Lipoproteins Link Enterococcus faecalis Virulence to Colitogenic Activity in IL-10-Deficient Mice Independent of Their Expression Levels.
Ocvirk S, Sava IG, Lengfelder I, Lagkouvardos I, Steck N, Roh JH, Tchaptchet S, Bao Y, Hansen JJ, Huebner J, Carroll IM, Murray BE, Sartor RB, Haller D. Ocvirk S, et al. PLoS Pathog. 2015 Jun 12;11(6):e1004911. doi: 10.1371/journal.ppat.1004911. eCollection 2015 Jun. PLoS Pathog. 2015. PMID: 26067254 Free PMC article.
References
- Xavier RJ, Podolsky DK. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448:427–434 - PubMed
- Chassaing B, Darfeuille-Michaud A. 2011. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 140:1720–1728 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical