Novel insights into the architecture and protein interaction network of yeast eIF3 - PubMed (original) (raw)
Novel insights into the architecture and protein interaction network of yeast eIF3
Sohail Khoshnevis et al. RNA. 2012 Dec.
Abstract
Translation initiation in eukaryotes is a multistep process requiring the orchestrated interaction of several eukaryotic initiation factors (eIFs). The largest of these factors, eIF3, forms the scaffold for other initiation factors, promoting their binding to the 40S ribosomal subunit. Biochemical and structural studies on eIF3 need highly pure eIF3. However, natively purified eIF3 comprise complexes containing other proteins such as eIF5. Therefore we have established in vitro reconstitution protocols for Saccharomyces cerevisiae eIF3 using its five recombinantly expressed and purified subunits. This reconstituted eIF3 complex (eIF3(rec)) exhibits the same size and activity as the natively purified eIF3 (eIF3(nat)). The homogeneity and stoichiometry of eIF3(rec) and eIF3(nat) were confirmed by analytical size exclusion chromatography, mass spectrometry, and multi-angle light scattering, demonstrating the presence of one copy of each subunit in the eIF3 complex. The reconstituted and native eIF3 complexes were compared by single-particle electron microscopy showing a high degree of structural conservation. The interaction network between eIF3 proteins was studied by means of limited proteolysis, analytical size exclusion chromatography, in vitro binding assays, and isothermal titration calorimetry, unveiling distinct protein domains and subcomplexes that are critical for the integrity of the protein network in yeast eIF3. Taken together, the data presented here provide a novel procedure to obtain highly pure yeast eIF3, suitable for biochemical and structural analysis, in addition to a detailed picture of the network of protein interactions within this complex.
Figures
FIGURE 1.
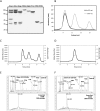
Reconstitution of yeast eIF3. (A) Comparison of reconstituted and native eIF3. Native eIF3 (3nat) copurifies with eIF5. Reconstituted eIF3 (3rec) comprises exclusively its five subunits. 3bcgi is a side product of the reconstitution of full eIF3 when Tif32 is added in lesser amounts compared with other subunits to promote the complex formation. All single recombinant subunits are shown on the gel to demonstrate the high degree of purity. (B) Size comparison between recombinant and native eIF3. Recombinant or native eIF3 were analyzed by size-exclusion chromatography (SEC). Recombinant eIF3 was prepared freshly by mixing five subunits in a nonstoichiometric ratio, therefore in addition to the peak for the full complex (between 9 and 10 mL), a second peak for Nip1-Prt1-Tif34-Tif35 complex (11–12 mL, corresponding to the third lane on A) and a third peak at ∼15 mL (corresponding to Tif34) were obtained. (C) Overlay of the SEC and MALS experiments for recombinant eIF3. The leftmost peak corresponds to recombinant eIF3. Molar mass distribution of the peak as calculated by MALS is indicated by a short line inside this peak and corresponds to a molecular weight of 340–354 kDa. The second peak corresponds to Nip1-Prt1-Tif34-Tif35 complex and corresponds to a molecular weight of 208–225 kDa. (D) Overlay of the SEC and MALS experiments for native eIF3. Molar mass distribution of the peak as calculated by MALS is indicated by a short line and corresponds to a molecular weight of 340–369 kDa. (E,F) Mass-spectrometry results for the eIF3rec (E) or eIF3nat (F). The upper panel is a zoom in the 10- to 120-kDa range of the spectrum for the control experiment without the cross-linker. The lower panel represents the whole spectrum for the cross-linked samples.
FIGURE 2.
Single particle EM analysis of native and recombinant eIF3 complexes. Selected 2D class averages of negatively stained samples of native eIF3 complex (upper row) and recombinant eIF3 complex (lower row) are shown.
FIGURE 3.
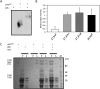
Activity tests for recombinant eIF3. (A) Analysis of the binding of eIF3 to the 40S ribosomal subunit by native PAGE and Western blot. A band shift toward higher molecular weights upon mixing 40S subunit and eIF3rec indicates the binding of eIF3 and 40S ribosome. (B) Luciferase assay performed with the cytoplasmic extracts of temperature-sensitive mutant cells treated under permissive (26°C) or nonpermissive (37°C) temperatures. To the cytoplasmic extracts, either recombinant (3rec) or native (3nat) eIF3 was added. In control experiments, only buffer was added. (C) SDS-PAGE analysis of cosedimentation experiment between eIF3rec and 40S. Mixtures were subjected to ultracentrifugation, and the total content of pellets (P) and supernatants (S) were loaded onto the gel. Under the conditions of this experiment, the majority of 40S subunits are found in the pellet, while all eIF3rec complexes remain in the supernatant. Upon mixing eIF3rec (0.1 μM) with 40S (1 μM), the factor was mainly found in the pellet, indicating binding to the 40S subunit. Addition of Hcr1 (1 μM) enhanced the binding of eIF3rec to the 40S. The leftmost and rightmost lanes show eIF3, which was used in this study and the marker, respectively.
FIGURE 4.
Limited proteolysis of the subcomplexes of recombinant eIF3. (A) Analytical SEC profile of the Prt1181C/Tif34/Tif35 complex digested with thermolysin after 24 h. Resulted fragments do not form the complex as manifested by the presence of two peaks shifted to the right (dark gray) compared to the noncleaved complex (light gray). (B) SDS-PAGE analysis of the SEC profile in panel A suggests the dissociation of the complex into a truncation of Prt1181C and Tif34 in complex with a truncation of Tif35. Numbers correspond to different fractions of the peak that are marked by a black bar on the chromatogram. 0 is the sample prior to gel filtration. (C) Analytical SEC of Tif32/Nip1 complex treated either with GluC for 2 h (dashed light gray) or with thermolysin for 1 d (solid light gray). Comparison with the noncleaved complex (solid dark gray) proposes the existence of only one complex in each case, indicating the preservation of the interactions between resulted fragments. (D) SDS-PAGE analysis of gel filtration runs in panel C shows the proteolytic fragments to be bound together. Left and right panels are different fractions of the peaks after GluC (dashed gray bar on the chromatogram) and thermolysin (solid gray bar on the chromatogram) treatments, respectively. In each case, 0 indicates the sample prior to the gel filtration. In all cases, M stands for molecular weight marker, and values are in kiloDaltons.
FIGURE 5.
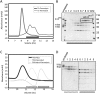
Prt1 and Tif35 interact weakly in vitro. (A) Pull-down interaction studies between Tif35, full-length Prt1, and Tif34. Lanes FT, W, and E refer to the flow-through of the Ni-NTA beads, their last wash step, and the elution of the protein. (B) Overlay of the analytical SEC profiles of Prt1181C (dashed black), Tif35 (solid black), and Prt1181C-Tif35 mixture (solid gray) shows a broadening of the peak in the case of the mixture without any considerable shift the retention volume compared with individual components. (C) SDS-PAGE analysis of the peak of the mixture of Prt1181C-Tif35 shows that it is in fact composed of two adjacent peaks (marked by two arrows) as would be judged by the broadening of the peak. (D) The equimolar interaction of Tif35 and Tif34 is driven by the release of heat, which compensates for the decrease in entropy. (E) Interaction of Tif34 with Prt1181C is also equimolar and enthalpy driven. In both cases, the upper panels show raw data of heat effect (in μcal/sec) of injections of Tif34 into 1.5 mL of Tif35 (D) or Prt1181C (E). The lower panels show the fitted binding isotherms. The data points were obtained by integration of heat signals plotted against the molar ratio of Tif34 to either of the interaction partners in the reaction cell. The solid lines represent calculated curves using the best fit parameters obtained by nonlinear least squares fitting.
FIGURE 6.
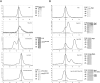
Mapping the interaction network of eIF3 by analytical SEC. (A) Full-length Prt1 interacts with the C-terminal region of Tif32. This interaction is not visible when the full-length Prt1 is replaced with a fragment starting from residue 181 (Prt1181C). (B) Nip1 interacts with Prt1-Tif34-Tif35 complex but not with each protein individually. Vertical lines are drawn to facilitate the comparison of the retention volumes of Nip1, Prt1, or Prt1181C in different experiments. In each case, the thick bar at the bottom of the graphs represents the fractions of the peak, which is resolved by SDS-PAGE.
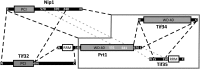
FIGURE 7.
Map of interactions between eIF3 subunits. The stable subcomplexes that could be separated by SEC are depicted by dashed lines. The interaction of Nip1 with Prt1-Tif34-Tif35 subcomplex is represented by a solid line around Prt1-Tif34-Tif35. Binding of Nip1370-570 to Prt1CTD was observed by Valasek et al. (2002), while the binding of Tif35CTD to Prt1CTD was concluded from indirect observations. Therefore these two interactions are represented by a different color (gray dashed lines).
References
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR 2007. Reconstitution of yeast translation initiation. Methods Enzymol 430: 111–145 - PubMed
- Algire MA, Maag D, Lorsch JR 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20: 251–262 - PubMed
- Asano K, Phan L, Anderson J, Hinnebusch AG 1998. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem 273: 18573–18585 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous