Prediction of protein phosphorylation sites by using the composition of k-spaced amino acid pairs - PubMed (original) (raw)
Prediction of protein phosphorylation sites by using the composition of k-spaced amino acid pairs
Xiaowei Zhao et al. PLoS One. 2012.
Abstract
As one of the most widespread protein post-translational modifications, phosphorylation is involved in many biological processes such as cell cycle, apoptosis. Identification of phosphorylated substrates and their corresponding sites will facilitate the understanding of the molecular mechanism of phosphorylation. Comparing with the labor-intensive and time-consuming experiment approaches, computational prediction of phosphorylation sites is much desirable due to their convenience and fast speed. In this paper, a new bioinformatics tool named CKSAAP_PhSite was developed that ignored the kinase information and only used the primary sequence information to predict protein phosphorylation sites. The highlight of CKSAAP_PhSite was to utilize the composition of k-spaced amino acid pairs as the encoding scheme, and then the support vector machine was used as the predictor. The performance of CKSAAP_PhSite was measured with a sensitivity of 84.81%, a specificity of 86.07% and an accuracy of 85.43% for serine, a sensitivity of 78.59%, a specificity of 82.26% and an accuracy of 80.31% for threonine as well as a sensitivity of 74.44%, a specificity of 78.03% and an accuracy of 76.21% for tyrosine. Experimental results obtained from cross validation and independent benchmark suggested that our method was very promising to predict phosphorylation sites and can be served as a useful supplement tool to the community. For public access, CKSAAP_PhSite is available at http://59.73.198.144/cksaap\_phsite/.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
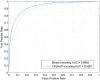
Figure 1. ROC curves of CKSAAP_PhSite and the binary encoding scheme in terms of serine (S) site prediction based on the training dataset.
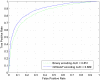
Figure 2. ROC curves of CKSAAP_PhSite and the binary encoding scheme in terms of threonine (T) site prediction based on the training dataset.
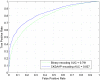
Figure 3. ROC curves of CKSAAP_PhSite and the binary encoding scheme in terms of tyrosine (Y) site prediction based on the training dataset.
Figure 4. Three Two-Sample-Logos of the position-specific residue composition surrounding the phosphorylated site and non-phosphorylated sites.
(A) serine site logo, (B) threonine site logo, (C) tyrosine site logo. These three logos were generated using the web server
and only residues significantly enriched and depleted surrounding phosphorylated sites (_t_-test, P<0.05) are shown.
Similar articles
- Prediction of mucin-type O-glycosylation sites in mammalian proteins using the composition of k-spaced amino acid pairs.
Chen YZ, Tang YR, Sheng ZY, Zhang Z. Chen YZ, et al. BMC Bioinformatics. 2008 Feb 18;9:101. doi: 10.1186/1471-2105-9-101. BMC Bioinformatics. 2008. PMID: 18282281 Free PMC article. - Prediction of ubiquitination sites by using the composition of k-spaced amino acid pairs.
Chen Z, Chen YZ, Wang XF, Wang C, Yan RX, Zhang Z. Chen Z, et al. PLoS One. 2011;6(7):e22930. doi: 10.1371/journal.pone.0022930. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829559 Free PMC article. - Prediction of lysine crotonylation sites by incorporating the composition of k-spaced amino acid pairs into Chou's general PseAAC.
Ju Z, He JJ. Ju Z, et al. J Mol Graph Model. 2017 Oct;77:200-204. doi: 10.1016/j.jmgm.2017.08.020. Epub 2017 Aug 24. J Mol Graph Model. 2017. PMID: 28886434 - A summary of computational resources for protein phosphorylation.
Xue Y, Gao X, Cao J, Liu Z, Jin C, Wen L, Yao X, Ren J. Xue Y, et al. Curr Protein Pept Sci. 2010 Sep;11(6):485-96. doi: 10.2174/138920310791824138. Curr Protein Pept Sci. 2010. PMID: 20491621 Review. - Recent Development of Machine Learning Methods in Microbial Phosphorylation Sites.
Rashid MM, Shatabda S, Hasan MM, Kurata H. Rashid MM, et al. Curr Genomics. 2020 Apr;21(3):194-203. doi: 10.2174/1389202921666200427210833. Curr Genomics. 2020. PMID: 33071613 Free PMC article. Review.
Cited by
- In silico functional and structural characterization of hepatitis B virus PreS/S-gene in Iranian patients infected with chronic hepatitis B virus genotype D.
Khodadad N, Seyedian SS, Moattari A, Biparva Haghighi S, Pirmoradi R, Abbasi S, Makvandi M. Khodadad N, et al. Heliyon. 2020 Jul 15;6(7):e04332. doi: 10.1016/j.heliyon.2020.e04332. eCollection 2020 Jul. Heliyon. 2020. PMID: 32695898 Free PMC article. - Rice_Phospho 1.0: a new rice-specific SVM predictor for protein phosphorylation sites.
Lin S, Song Q, Tao H, Wang W, Wan W, Huang J, Xu C, Chebii V, Kitony J, Que S, Harrison A, He H. Lin S, et al. Sci Rep. 2015 Jul 7;5:11940. doi: 10.1038/srep11940. Sci Rep. 2015. PMID: 26149854 Free PMC article. - PTM-ssMP: A Web Server for Predicting Different Types of Post-translational Modification Sites Using Novel Site-specific Modification Profile.
Liu Y, Wang M, Xi J, Luo F, Li A. Liu Y, et al. Int J Biol Sci. 2018 May 22;14(8):946-956. doi: 10.7150/ijbs.24121. eCollection 2018. Int J Biol Sci. 2018. PMID: 29989096 Free PMC article. - An ensemble method approach to investigate kinase-specific phosphorylation sites.
Datta S, Mukhopadhyay S. Datta S, et al. Int J Nanomedicine. 2014 May 10;9:2225-39. doi: 10.2147/IJN.S57526. eCollection 2014. Int J Nanomedicine. 2014. PMID: 24872686 Free PMC article. - Precise Prediction of Calpain Cleavage Sites and Their Aberrance Caused by Mutations in Cancer.
Liu ZX, Yu K, Dong J, Zhao L, Liu Z, Zhang Q, Li S, Du Y, Cheng H. Liu ZX, et al. Front Genet. 2019 Aug 8;10:715. doi: 10.3389/fgene.2019.00715. eCollection 2019. Front Genet. 2019. PMID: 31440276 Free PMC article.
References
- Uddin S, Lekmine F, Sassano A, Rui H, Fish EN, Platanias LC (2003) Role of Stat5 in type I interferon-signaling and transcriptional regulation. Biochem Biophys Res Commun 308: 325–330. - PubMed
- Bu YH, He YL, Zhou HD, Liu W, Peng D, Tang AG, Tang LL, Xie H, Huang QX, Luo XH, Liao EY (2010) Insulin receptor substrate 1 regulates the cellular differentiation and the matrix metallopeptidase expression of preosteoblastic cells. J Endocrinol 206: 271–277. - PubMed
- Kim SH, Lee CE (2011) Counter-regulation mechanism of IL-4 and IFN-α signal transduction through cytosolic retention of the pY-STAT6: pY-STAT2:p48 complex. Eur J Immunol 41: 461–472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This research is partially supported by the National Natural Science Foundation of China under Grant Nos. 60803102, 61070084, and also funded by the Natural Science Foundation of Jilin Province (Nos. 20101506 and 20110104). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Research Materials