Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes - PubMed (original) (raw)
Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes
Rolf S Kaas et al. BMC Genomics. 2012.
Abstract
Background: Escherichia coli exists in commensal and pathogenic forms. By measuring the variation of individual genes across more than a hundred sequenced genomes, gene variation can be studied in detail, including the number of mutations found for any given gene. This knowledge will be useful for creating better phylogenies, for determination of molecular clocks and for improved typing techniques.
Results: We find 3,051 gene clusters/families present in at least 95% of the genomes and 1,702 gene clusters present in 100% of the genomes. The former 'soft core' of about 3,000 gene families is perhaps more biologically relevant, especially considering that many of these genome sequences are draft quality. The E. coli pan-genome for this set of isolates contains 16,373 gene clusters.A core-gene tree, based on alignment and a pan-genome tree based on gene presence/absence, maps the relatedness of the 186 sequenced E. coli genomes. The core-gene tree displays high confidence and divides the E. coli strains into the observed MLST type clades and also separates defined phylotypes.
Conclusion: The results of comparing a large and diverse E. coli dataset support the theory that reliable and good resolution phylogenies can be inferred from the core-genome. The results further suggest that the resolution at the isolate level may, subsequently be improved by targeting more variable genes. The use of whole genome sequencing will make it possible to eliminate, or at least reduce, the need for several typing steps used in traditional epidemiology.
Figures
Figure 1
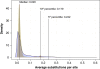
Progress of Homolog Gene Cluster calculation as each genome is added. Two circles exist (red & blue) for each genome added from genome no. 9 up to and including genome no. 186. Red represents the number of core HGCs after the addition of a genome and blue represents the number of pan HGCs after the addition of a genome.
Figure 2
HGC Variation plot. A Density plot was created from the calculation of nucleotide diversity within each HGC. The blue plot was created from all the HGCs. The red plot only includes the strict core HGCs. The green plot includes the soft core (95%) HGCs. Intersection between core plots is yellow.
Figure 3
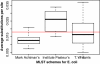
Box plot of MLST gene variation. A box plot presenting the distribution of nucleotide diversity within each of the three MLST schemes. The red line represents the median of percent identity for HGCs in the core (~0.018 substitutions per site).
Figure 4
Core-gene tree close-up on O157:H7 strains. The tree is a close-up of the O157:H7 clade from the core-gene tree presented in Figure 6. The names has been colored according to the three outbreaks described in [21]. Blue strains represent the spinach outbreak, red strains represent the Taco Bell outbreak and the green strains represent the Taco John outbreak. Branch lengths have been modified to create the best visual output and thus have no value.
Figure 5
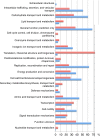
General function of conserved and variable HGCs. The difference in functional annotations between conserved and variable HGCs. Conserved here defined as the quarter of HGCs with the lowest nucleotide diversity (red bars) and variable defined as the quarter of HGCs with the highest nucleotide diversity (blue bars). Each HGC has a functional profile. A functional profile consists of one or more functional categories. The bars represent the percentage of HGC profiles, which contain the functional category listed to the immediate left of the bars.
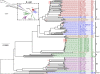
Figure 6
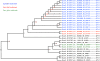
Core-gene tree. The E. coli tree was created from the alignment of 1,278 core-genes from the 186 E. coli genomes. MLST types are annotated to the far right of each genome name. The Escherichia genus tree was created from 297 core-genes. The phylotypes, as determined by the in silico Clermont [15] method, are marked with the colors blue (A), red (B1), purple (B2), green (D), and the Shigella genomes are marked with the color brown. At each node a black circle indicates a bootstrap value of 1, a grey circle a bootstrap value between 1 and 0.7 and a red number indicate an actual bootstrap value below 0.7. The dashed line in the figure represents a branch, which has been manually shortened by the authors to fit the figure on a printed page. The original tree with all bootstrap values can be seen in Additional file 2. Both trees are unrooted, but the E. coli tree has been visually rooted on the node leading to Clade I.
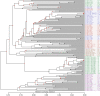
Figure 7
Pan genome tree. The tree was created based on the presence or absence of 16,373 HGCs in the 186 E. coli genomes. MLST types are annotated to the far right of each genome name. The phylotypes are marked with the colors blue (A), red (B1), purple (B2), green (D), and the Shigella genomes are marked with the color brown. Bootstrap values are annotated at each node as a percentage between 0 and 100. At each node a black circle indicates a bootstrap value of 100, a grey circle indicates a bootstrap value between 100 and 70 and a red circle indicates a bootstrap value below 70. The original tree with all bootstrap values can be seen in Additional file 3 .
Similar articles
- A phylogenomic analysis of Escherichia coli / Shigella group: implications of genomic features associated with pathogenicity and ecological adaptation.
Zhang Y, Lin K. Zhang Y, et al. BMC Evol Biol. 2012 Sep 7;12:174. doi: 10.1186/1471-2148-12-174. BMC Evol Biol. 2012. PMID: 22958895 Free PMC article. - Phylomark, a tool to identify conserved phylogenetic markers from whole-genome alignments.
Sahl JW, Matalka MN, Rasko DA. Sahl JW, et al. Appl Environ Microbiol. 2012 Jul;78(14):4884-92. doi: 10.1128/AEM.00929-12. Epub 2012 May 11. Appl Environ Microbiol. 2012. PMID: 22582056 Free PMC article. - Comparative genomics of European avian pathogenic E. Coli (APEC).
Cordoni G, Woodward MJ, Wu H, Alanazi M, Wallis T, La Ragione RM. Cordoni G, et al. BMC Genomics. 2016 Nov 22;17(1):960. doi: 10.1186/s12864-016-3289-7. BMC Genomics. 2016. PMID: 27875980 Free PMC article. - Comparison of 61 sequenced Escherichia coli genomes.
Lukjancenko O, Wassenaar TM, Ussery DW. Lukjancenko O, et al. Microb Ecol. 2010 Nov;60(4):708-20. doi: 10.1007/s00248-010-9717-3. Epub 2010 Jul 11. Microb Ecol. 2010. PMID: 20623278 Free PMC article. Review. - Are Escherichia coli Pathotypes Still Relevant in the Era of Whole-Genome Sequencing?
Robins-Browne RM, Holt KE, Ingle DJ, Hocking DM, Yang J, Tauschek M. Robins-Browne RM, et al. Front Cell Infect Microbiol. 2016 Nov 18;6:141. doi: 10.3389/fcimb.2016.00141. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27917373 Free PMC article. Review.
Cited by
- Whole genome sequence analysis reveals high genomic diversity and potential host-driven adaptations among multidrug-resistant Escherichia coli from pre-weaned dairy calves.
Lee KY, Schlesener CL, Aly SS, Huang BC, Li X, Atwill ER, Weimer BC. Lee KY, et al. Front Microbiol. 2024 Sep 3;15:1420300. doi: 10.3389/fmicb.2024.1420300. eCollection 2024. Front Microbiol. 2024. PMID: 39296303 Free PMC article. - Corynebacterium pseudotuberculosis: Whole genome sequencing reveals unforeseen and relevant genetic diversity in this pathogen.
Hiller E, Hörz V, Sting R. Hiller E, et al. PLoS One. 2024 Aug 26;19(8):e0309282. doi: 10.1371/journal.pone.0309282. eCollection 2024. PLoS One. 2024. PMID: 39186721 Free PMC article. - Synonymous rpsH variants: the common denominator in Escherichia coli adapting to ionizing radiation.
Stemwedel K, Haase N, Christ S, Bogdanova NV, Rudorf S. Stemwedel K, et al. NAR Genom Bioinform. 2024 Aug 24;6(3):lqae110. doi: 10.1093/nargab/lqae110. eCollection 2024 Sep. NAR Genom Bioinform. 2024. PMID: 39184377 Free PMC article. - Patterns of Change in Nucleotide Diversity Over Gene Length.
Ali F. Ali F. Genome Biol Evol. 2024 Apr 2;16(4):evae078. doi: 10.1093/gbe/evae078. Genome Biol Evol. 2024. PMID: 38608148 Free PMC article. - Gene presence/absence variation in Mytilus galloprovincialis and its implications in gene expression and adaptation.
Saco A, Rey-Campos M, Gallardo-Escárate C, Gerdol M, Novoa B, Figueras A. Saco A, et al. iScience. 2023 Sep 4;26(10):107827. doi: 10.1016/j.isci.2023.107827. eCollection 2023 Oct 20. iScience. 2023. PMID: 37744033 Free PMC article.
References
- Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJM, Brinkman FSL, Brunham RC, Tang P. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–739. doi: 10.1056/NEJMoa1003176. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources