Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution - PubMed (original) (raw)
Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution
Vuong Tran et al. Science. 2012.
Abstract
Stem cells can self-renew and generate differentiating daughter cells. It is not known whether these cells maintain their epigenetic information during asymmetric division. Using a dual-color method to differentially label "old" versus "new" histones in Drosophila male germline stem cells (GSCs), we show that preexisting canonical H3, but not variant H3.3, histones are selectively segregated to the GSC, whereas newly synthesized histones incorporated during DNA replication are enriched in the differentiating daughter cell. The asymmetric histone distribution occurs in GSCs but not in symmetrically dividing progenitor cells. Furthermore, if GSCs are genetically manipulated to divide symmetrically, this asymmetric mode is lost. This work suggests that stem cells retain preexisting canonical histones during asymmetric cell divisions, probably as a mechanism to maintain their unique molecular properties.
Figures
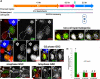
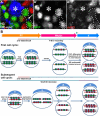
Figure 1. Experimental design and potential results
(A) A diagram of the GSC niche. HUB-hub cells, CySC- cyst progenitor/somatic stem cell. (B) Immunofluorescent image of the niche: HUB (anti-Fas III, red, asterisk), GSC-GB pair expressing H3-GFP (green, dotted outline) connected by a spectrosome (anti-α-Spectrin, red, arrow). (C) The UASp-FRT-histone-GFP-PolyA-FRT-histone-mKO-PolyA transgene. UAS:
u
pstream
a
ctivating
s
equence;
F
RT: FLP (flippase)
r
ecombination
t
arget; histone: H3, H2B, or histone variant H3.3. nanos-Gal4: a germline-specific driver. hs-FLP: the yeast FLP recombinase controlled by the
h
eat
s
hock (hs) promoter. (D–E) Two potential results: for simplicity, only one GSC-GB pair is shown, and each entire cell is colored according to histone fluorescence.
Figure 2. H3 is asymmetrically segregated during the second GSC division after heat shock
(A) Heat shock regime. H3 is distributed asymmetrically in GSC vs. GB (B-B”) but symmetrically in two-cell spermatogonia (C-C”). (D-D”) H3.3 is distributed symmetrically in GSC vs. GB. H3 distribution pattern in GSCs: (E-E”) G2 phase, (F-F”) anaphase, (G-G”) telophase. Scale: 5μm. Asterisk: HUB (anti-FasIII); arrow: spectrosome (anti-α spectrin). (H) Quantification of GFP and mKO fluorescence intensity ratio (Table S2). H3 GSC/GB GFP ratio > 1 (* P<10−4), GSC/GB mKO ratio < 1 (* P<10−4), _N_=15. H3 two-cell spermatogonial (SG) SG1/SG2 GFP ratio (# P=0.103) and mKO ratio (# P=0.684) insignificantly different from 1, _N_=16. H3.3 GSC/GB GFP ratio (# P=0.513) and mKO ratio (# P=0.532) insignificantly different from 1, _N_=12. Error bars: S.E. P-value: one-sample t-test.
Figure 3. H3 is asymmetrically distributed after the first GSC division after heat shock
(A) Heat shock regime. H3 is distributed asymmetrically in GSC vs. GB (B-B”) but symmetrically in two-cell spermatogonia (C-C”). (D-D”) H3.3 is distributed symmetrically in GSC vs. GB. (E-E”) A telophase GSC. Scale: 5μm. Asterisk: HUB (anti-FasIII); arrow: spectrosome (anti-α spectrin). (F) Quantification of GFP and mKO fluorescence intensity ratio (Table S4). H3 GSC/GB GFP ratio > 1 (* P< 10−4), GSC/GB mKO ratio < 1 (* P< 10−4), _N_=12. H3 two-cell spermatogonial (SG) SG1/SG2 GFP ratio (# P=0.225) and mKO ratio (# P=0.365) insignificantly different from 1, _N_=11. H3.3 GSC/GB GFP ratio (# P=0.970) and mKO ratio (# P=0.594) insignificantly different from 1, _N_=13. Error bars: S.E. P-value: one-sample t-test.
Figure 4. Loss of asymmetric H3 distribution pattern upon overexpression of upd
(A-A”) H3-GFP (A') and H3-mKO (A”) in nanos-Gal4; UAS-upd testis. Asterisk: HUB (anti-FasIII).
Similar articles
- Asymmetric distribution of histones during Drosophila male germline stem cell asymmetric divisions.
Tran V, Feng L, Chen X. Tran V, et al. Chromosome Res. 2013 May;21(3):255-69. doi: 10.1007/s10577-013-9356-x. Chromosome Res. 2013. PMID: 23681658 Free PMC article. Review. - Asymmetric Histone Inheritance in Asymmetrically Dividing Stem Cells.
Wooten M, Ranjan R, Chen X. Wooten M, et al. Trends Genet. 2020 Jan;36(1):30-43. doi: 10.1016/j.tig.2019.10.004. Epub 2019 Nov 18. Trends Genet. 2020. PMID: 31753528 Free PMC article. Review. - A single N-terminal amino acid determines the distinct roles of histones H3 and H3.3 in the Drosophila male germline stem cell lineage.
Chandrasekhara C, Ranjan R, Urban JA, Davis BEM, Ku WL, Snedeker J, Zhao K, Chen X. Chandrasekhara C, et al. PLoS Biol. 2023 May 1;21(5):e3002098. doi: 10.1371/journal.pbio.3002098. eCollection 2023 May. PLoS Biol. 2023. PMID: 37126497 Free PMC article. - Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline.
Xie J, Wooten M, Tran V, Chen BC, Pozmanter C, Simbolon C, Betzig E, Chen X. Xie J, et al. Cell. 2015 Nov 5;163(4):920-33. doi: 10.1016/j.cell.2015.10.002. Epub 2015 Oct 29. Cell. 2015. PMID: 26522592 Free PMC article. - Live imaging of the Drosophila ovarian niche shows spectrosome and centrosome dynamics during asymmetric germline stem cell division.
Villa-Fombuena G, Lobo-Pecellín M, Marín-Menguiano M, Rojas-Ríos P, González-Reyes A. Villa-Fombuena G, et al. Development. 2021 Sep 15;148(18):dev199716. doi: 10.1242/dev.199716. Epub 2021 Sep 17. Development. 2021. PMID: 34370012 Free PMC article.
Cited by
- What do you mean, "epigenetic"?
Deans C, Maggert KA. Deans C, et al. Genetics. 2015 Apr;199(4):887-96. doi: 10.1534/genetics.114.173492. Genetics. 2015. PMID: 25855649 Free PMC article. - Breaking Symmetry: The Asymmetries in Epigenetic Inheritance.
Zion E, Chen X. Zion E, et al. Biochem (Lond). 2021 Feb;43(1):14-19. doi: 10.1042/bio_2020_110. Epub 2021 Jan 29. Biochem (Lond). 2021. PMID: 34354328 Free PMC article. - Histone variants on the move: substrates for chromatin dynamics.
Talbert PB, Henikoff S. Talbert PB, et al. Nat Rev Mol Cell Biol. 2017 Feb;18(2):115-126. doi: 10.1038/nrm.2016.148. Epub 2016 Dec 7. Nat Rev Mol Cell Biol. 2017. PMID: 27924075 Review. - The Inherent Asymmetry of DNA Replication.
Snedeker J, Wooten M, Chen X. Snedeker J, et al. Annu Rev Cell Dev Biol. 2017 Oct 6;33:291-318. doi: 10.1146/annurev-cellbio-100616-060447. Epub 2017 Aug 11. Annu Rev Cell Dev Biol. 2017. PMID: 28800257 Free PMC article. Review. - Characterization of histone inheritance patterns in the Drosophila female germline.
Kahney EW, Zion EH, Sohn L, Viets-Layng K, Johnston R, Chen X. Kahney EW, et al. EMBO Rep. 2021 Jul 5;22(7):e51530. doi: 10.15252/embr.202051530. Epub 2021 May 25. EMBO Rep. 2021. PMID: 34031963 Free PMC article.
References
- Ringrose L, Paro R. Annu Rev Genet. 2004;38:413. - PubMed
- Jacobs JJ, van Lohuizen M. Biochim Biophys Acta. 2002 Jun 21;1602:151. - PubMed
- Turner BM. Cell. 2002 Nov 1;111:285. - PubMed
- Knoblich JA. Cell. 2008 Feb 22;132:583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007231/GM/NIGMS NIH HHS/United States
- R01 HD065816/HD/NICHD NIH HHS/United States
- R01HD065816/HD/NICHD NIH HHS/United States
- R21 HD065089/HD/NICHD NIH HHS/United States
- R21HD065089/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials