Rfam 11.0: 10 years of RNA families - PubMed (original) (raw)
. 2013 Jan;41(Database issue):D226-32.
doi: 10.1093/nar/gks1005. Epub 2012 Nov 3.
Affiliations
- PMID: 23125362
- PMCID: PMC3531072
- DOI: 10.1093/nar/gks1005
Rfam 11.0: 10 years of RNA families
Sarah W Burge et al. Nucleic Acids Res. 2013 Jan.
Abstract
The Rfam database (available via the website at http://rfam.sanger.ac.uk and through our mirror at http://rfam.janelia.org) is a collection of non-coding RNA families, primarily RNAs with a conserved RNA secondary structure, including both RNA genes and mRNA cis-regulatory elements. Each family is represented by a multiple sequence alignment, predicted secondary structure and covariance model. Here we discuss updates to the database in the latest release, Rfam 11.0, including the introduction of genome-based alignments for large families, the introduction of the Rfam Biomart as well as other user interface improvements. Rfam is available under the Creative Commons Zero license.
Figures
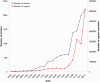
Figure 1.
Growth of Rfam families and sequence regions annotated per each Rfam release. Dates are in MM/YY format.
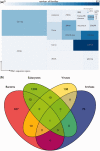
Figure 2.
(a) Rfam types organized according to their coverage of sequence space. Size of rectangles is proportional to the number of regions annotated by families of that Rfam type; colour of rectangles is proportional to the number of families of that type. (b) Taxonomic coverage of Rfam families. Families have been categorized according to the taxonomic groups covered by the seed sequences, and families in clans are treated as belonging to a single family. This analysis omits six families where the seed contains only unclassified sequences.
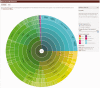
Figure 3.
Sunburst visualization of family taxonomy for RF01051, the cyclic di-GMP-I riboswitch. Users may select regions of taxonomic space and use the controls on the left to download an alignment of their chosen subset of species.
Similar articles
- Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families.
Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. Kalvari I, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D335-D342. doi: 10.1093/nar/gkx1038. Nucleic Acids Res. 2018. PMID: 29112718 Free PMC article. - Rfam: an RNA family database.
Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Griffiths-Jones S, et al. Nucleic Acids Res. 2003 Jan 1;31(1):439-41. doi: 10.1093/nar/gkg006. Nucleic Acids Res. 2003. PMID: 12520045 Free PMC article. - Rfam 12.0: updates to the RNA families database.
Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, Finn RD. Nawrocki EP, et al. Nucleic Acids Res. 2015 Jan;43(Database issue):D130-7. doi: 10.1093/nar/gku1063. Epub 2014 Nov 11. Nucleic Acids Res. 2015. PMID: 25392425 Free PMC article. - Customized strategies for discovering distant ncRNA homologs.
Mosig A, Zhu L, Stadler PF. Mosig A, et al. Brief Funct Genomic Proteomic. 2009 Nov;8(6):451-60. doi: 10.1093/bfgp/elp035. Epub 2009 Sep 24. Brief Funct Genomic Proteomic. 2009. PMID: 19779009 Review. - An Ariadne's thread to the identification and annotation of noncoding RNAs in eukaryotes.
Soldà G, Makunin IV, Sezerman OU, Corradin A, Corti G, Guffanti A. Soldà G, et al. Brief Bioinform. 2009 Sep;10(5):475-89. doi: 10.1093/bib/bbp022. Epub 2009 Apr 21. Brief Bioinform. 2009. PMID: 19383843 Review.
Cited by
- antaRNA: ant colony-based RNA sequence design.
Kleinkauf R, Mann M, Backofen R. Kleinkauf R, et al. Bioinformatics. 2015 Oct 1;31(19):3114-21. doi: 10.1093/bioinformatics/btv319. Epub 2015 May 27. Bioinformatics. 2015. PMID: 26023105 Free PMC article. - De novo assembly of mud loach (Misgurnus anguillicaudatus) skin transcriptome to identify putative genes involved in immunity and epidermal mucus secretion.
Long Y, Li Q, Zhou B, Song G, Li T, Cui Z. Long Y, et al. PLoS One. 2013;8(2):e56998. doi: 10.1371/journal.pone.0056998. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437293 Free PMC article. - The Ordospora colligata genome: Evolution of extreme reduction in microsporidia and host-to-parasite horizontal gene transfer.
Pombert JF, Haag KL, Beidas S, Ebert D, Keeling PJ. Pombert JF, et al. mBio. 2015 Jan 13;6(1):e02400-14. doi: 10.1128/mBio.02400-14. mBio. 2015. PMID: 25587016 Free PMC article. - Habitat visualization and genomic analysis of "Candidatus Pantoea carbekii," the primary symbiont of the brown marmorated stink bug.
Kenyon LJ, Meulia T, Sabree ZL. Kenyon LJ, et al. Genome Biol Evol. 2015 Jan 12;7(2):620-35. doi: 10.1093/gbe/evv006. Genome Biol Evol. 2015. PMID: 25587021 Free PMC article. - Extracellular and Intracellular Lanthanide Accumulation in the Methylotrophic Beijerinckiaceae Bacterium RH AL1.
Wegner CE, Westermann M, Steiniger F, Gorniak L, Budhraja R, Adrian L, Küsel K. Wegner CE, et al. Appl Environ Microbiol. 2021 Jun 11;87(13):e0314420. doi: 10.1128/AEM.03144-20. Epub 2021 Jun 11. Appl Environ Microbiol. 2021. PMID: 33893117 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources