Visualizing a complete Siphoviridae member by single-particle electron microscopy: the structure of lactococcal phage TP901-1 - PubMed (original) (raw)
Visualizing a complete Siphoviridae member by single-particle electron microscopy: the structure of lactococcal phage TP901-1
Cecilia Bebeacua et al. J Virol. 2013 Jan.
Abstract
Tailed phages are genome delivery machines exhibiting unequaled efficiency acquired over more than 3 billion years of evolution. Siphophages from the P335 and 936 families infect the Gram-positive bacterium Lactococcus lactis using receptor-binding proteins anchored to the host adsorption apparatus (baseplate). Crystallographic and electron microscopy (EM) studies have shed light on the distinct adsorption strategies used by phages of these two families, suggesting that they might also rely on different infection mechanisms. Here, we report electron microscopy reconstructions of the whole phage TP901-1 (P335 species) and propose a composite EM model of this gigantic molecular machine. Our results suggest conservation of structural proteins among tailed phages and add to the growing body of evidence pointing to a common evolutionary origin for these virions. Finally, we propose that host adsorption apparatus architectures have evolved in correlation with the nature of the receptors used during infection.
Figures
Fig 1
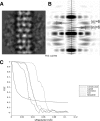
EM parameters of TP901-1 structure. (A and B) Determination of the helical parameters of the TP901 tail. An average of aligned tail tube segments (A) was used to generate the Fourier transform (B). The meridional line is marked by a dotted line. The layer lines are marked by arrows that also indicate their Bessel orders (6 and −6). This indexation showed the 6-fold rotational symmetry of the TP901 helical tail. (C) Fourier shell correlation (FSC) curves of the final three-dimensional (3D) reconstructions obtained by correlation of two different 3D reconstructions created by splitting the particle set into two subsets. The resolution was estimated by the 1/2-bit cutoff threshold criterion as 15 Å for the capsid, 21 Å for the connector, 21 Å for the helical tail, and 25 Å for the baseplate.
Fig 2
The 15-Å-resolution cryo-EM reconstruction of the TP901-1 mature capsid. (A) Surface rendering of the icosahedral reconstruction low-pass filtered at 15 Å viewed along an icosahedral 2-fold axis. The capsid measures 660 Å along its 5-fold axis. (B) Cross section of the capsid reconstruction showing the layers of the dsDNA genome organized as concentric shells. (C) Pseudoatomic model of the TP901-1 mature capsid fitted in the reconstruction.
Fig 3
The 20-Å-resolution reconstruction of the TP901-1 connector. (A to C) The connector occupies a unique capsid vertex. (A) Side view. (B) View from the distal extremity along the tail tube. (C) Cross section of the capsid showing the portal protruding into it. (D) The SPP1 portal and the first head completion protein dodecamer (SPP1 gp15) are fitted into the connector reconstruction. Note the additional density surrounding the gp15 ring that likely corresponds to the collar/whiskers. The stopper (equivalent to SPP1 gp16) and the tail terminator are postulated to account for the remaining density. (E) View along the tail axis showing the fitting of the SPP1 portal dodecamer into the corresponding TP901-1 EM density (the region corresponding to the capsid density has been computationally removed for clarity). (F) View along the tail axis showing the fitting of the SPP1 first head completion protein into the corresponding TP901-1 EM density.
Fig 4
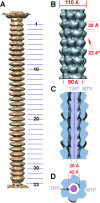
The 20-Å-resolution reconstruction of the TP901-1 tail. (A) Sixfold-averaged reconstruction of the TP901-1 phage tail from a few selected virions exhibiting an almost straight tail, making it possible to obtain the number of MTPs. (B) Detailed view of the reconstruction of a segment of the tail (7 MTP rings) using helical symmetry. The helical parameters of the tail are shown. (C) Cross section of the tail segment (blue) along its long axis revealing the internal TMP (violet). (D) Cross section of the tail segment orthogonal to its long axis (the color scheme is the same as in panel C).
Fig 5
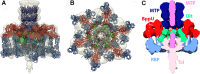
The 20-Å-resolution reconstruction of the TP901-1 distal tail region (the baseplate). The TP901-1 baseplate crystal structure was rigid-body fitted in the reconstruction. The color code is as follows: Dit (green), BppU (red), and BppL (light blue). The MTP hexamers were modeled using the Hcp type VI secretion system protein (dark blue). The N-terminal region of the tail fiber was modeled using the phage p2 ORF16 in closed conformation (pink). (A) Side view. (B) View along the tail axis from the distal extremity toward the capsid. (C) Cross section of the baseplate EM map. The assignment of the EM density to the different ORFs was performed using the X-ray structure.
Fig 6
Assembled complete structure of the TP901-1 phage. The complete phage was assembled by fitting the individually refined reconstructions into the map obtained for the full phage.
Similar articles
- Cryo-electron microscopy structure of lactococcal siphophage 1358 virion.
Spinelli S, Bebeacua C, Orlov I, Tremblay D, Klaholz BP, Moineau S, Cambillau C. Spinelli S, et al. J Virol. 2014 Aug;88(16):8900-10. doi: 10.1128/JVI.01040-14. Epub 2014 May 28. J Virol. 2014. PMID: 24872584 Free PMC article. - The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Legrand P, Collins B, Blangy S, Murphy J, Spinelli S, Gutierrez C, Richet N, Kellenberger C, Desmyter A, Mahony J, van Sinderen D, Cambillau C. Legrand P, et al. mBio. 2016 Jan 26;7(1):e01781-15. doi: 10.1128/mBio.01781-15. mBio. 2016. PMID: 26814179 Free PMC article. - Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009.
Vegge CS, Vogensen FK, Mc Grath S, Neve H, van Sinderen D, Brøndsted L. Vegge CS, et al. J Bacteriol. 2006 Jan;188(1):55-63. doi: 10.1128/JB.188.1.55-63.2006. J Bacteriol. 2006. PMID: 16352821 Free PMC article. - Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages.
Dunne M, Hupfeld M, Klumpp J, Loessner MJ. Dunne M, et al. Viruses. 2018 Jul 28;10(8):397. doi: 10.3390/v10080397. Viruses. 2018. PMID: 30060549 Free PMC article. Review. - Structures and host-adhesion mechanisms of lactococcal siphophages.
Spinelli S, Veesler D, Bebeacua C, Cambillau C. Spinelli S, et al. Front Microbiol. 2014 Jan 16;5:3. doi: 10.3389/fmicb.2014.00003. eCollection 2014. Front Microbiol. 2014. PMID: 24474948 Free PMC article. Review.
Cited by
- Ubiquitous Carbohydrate Binding Modules Decorate 936 Lactococcal Siphophage Virions.
Hayes S, Mahony J, Vincentelli R, Ramond L, Nauta A, van Sinderen D, Cambillau C. Hayes S, et al. Viruses. 2019 Jul 9;11(7):631. doi: 10.3390/v11070631. Viruses. 2019. PMID: 31324000 Free PMC article. - The Francisella Type VI Secretion System.
Clemens DL, Lee BY, Horwitz MA. Clemens DL, et al. Front Cell Infect Microbiol. 2018 Apr 23;8:121. doi: 10.3389/fcimb.2018.00121. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29740542 Free PMC article. Review. - Molecular architecture of tailed double-stranded DNA phages.
Fokine A, Rossmann MG. Fokine A, et al. Bacteriophage. 2014 Jan 1;4(1):e28281. doi: 10.4161/bact.28281. Epub 2014 Feb 21. Bacteriophage. 2014. PMID: 24616838 Free PMC article. Review. - Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein.
Farenc C, Spinelli S, Vinogradov E, Tremblay D, Blangy S, Sadovskaya I, Moineau S, Cambillau C. Farenc C, et al. J Virol. 2014 Jun;88(12):7005-15. doi: 10.1128/JVI.00739-14. Epub 2014 Apr 9. J Virol. 2014. PMID: 24719416 Free PMC article. - Siphophage 0105phi7-2 of Bacillus thuringiensis: Novel Propagation, DNA, and Genome-Implied Assembly.
Roberts SM, Aldis M, Wright ET, Gonzales CB, Lai Z, Weintraub ST, Hardies SC, Serwer P. Roberts SM, et al. Int J Mol Sci. 2023 May 18;24(10):8941. doi: 10.3390/ijms24108941. Int J Mol Sci. 2023. PMID: 37240285 Free PMC article.
References
- Brussow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108:13–16 - PubMed
- Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, Mesyanzhinov VV, Rossmann MG. 2003. Three-dimensional structure of bacteriophage T4 baseplate. Nat. Struct. Biol. 10:688–693 - PubMed
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. 2006. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312:1791–1795 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources