Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel - PubMed (original) (raw)
. 2012 Dec;19(12):1234-41.
doi: 10.1038/nsmb.2438. Epub 2012 Nov 11.
Affiliations
- PMID: 23142978
- PMCID: PMC3678077
- DOI: 10.1038/nsmb.2438
Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel
Bettina Bradatsch et al. Nat Struct Mol Biol. 2012 Dec.
Abstract
Preribosomal particles evolve in the nucleus through transient interaction with biogenesis factors before export to the cytoplasm. Here, we report the architecture of the late pre-60S particle, purified from Saccharomyces cerevisiae, through Arx1, a nuclear export factor with structural homology to methionine aminopeptidases, or its binding partner Alb1. Cryo-EM reconstruction of the Arx1 particle at 11.9-Å resolution reveals regions of extra density on the pre-60S particle attributed to associated biogenesis factors, confirming the immature state of the nascent subunit. One of these densities could be unambiguously assigned to Arx1. Immunoelectron microscopy and UV cross-linking localize Arx1 close to the ribosomal exit tunnel, in direct contact with ES27, a highly dynamic eukaryotic rRNA expansion segment. The binding of Arx1 at the exit tunnel may position this export factor to prevent premature recruitment of ribosome-associated factors active during translation.
Figures
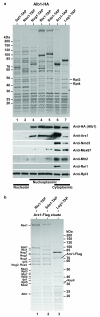
Figure 1. Arx1 and Alb1 are present on the same pre-60S particles
(a) Tandem affinity purification of different pre-60S particles (nucleolar, nucleoplasmic, cytoplasmic) using the indicated TAP-tagged bait proteins from the respective yeast strains harboring chromosomally integrated Alb1-HA. The final eluates of these purifications were analyzed by SDS-PAGE and Coomassie staining (upper panel), or Western blotting (lower panel) using the indicated antibodies against pre-ribosomal factors and r-proteins. The position of the respective Coomassie-stained TAP-tagged bait protein is marked by a star. A protein marker is indicated on the left, the position of two r-proteins (Rpl3, Rpl4) on the right, and the subcellular location of the purified particles below. (b) Split-tag affinity purification reveals Arx1 as a stoichiometric factor both on nucleoplasmic and cytoplasmic pre-60S particles. Arx1-containing pre-60S particles were affinity-purified via the split-tag method using the indicated Rix1-TAP, Sda1-TAP and Lsg1-TAP bait proteins with integrated Arx1-Flag. It was first purified on IgG-beads exploiting the ProtA-tag, followed by purification of the TEV-eluate on Flag-beads using co-enriched Arx1-Flag as affinity-ligand. The final eluates obtained after Flag peptide elution were analyzed by SDS-PAGE and Coomassie staining. Indicated are the position of Arx1-Flag, Alb1 and other prominent biogenesis factors on the right and left, respectively, the TAP-tagged bait proteins by a star, and a protein marker on the right.
Figure 2. Cryo-EM reconstruction of the Arx1-particle
(a) Cryo-EM reconstruction of the Alb1-TAP affinity-purified pre-60S at 11.9 Å resolution (right) in comparison to mature 60S subunit, isolated from S. cerevisiae RNC-Ssh1 (left). Labeled are: L1-stalk (L1), central protuberance (CP), stalk base (SB), P-stalk (St), exit tunnel (ET). The pre-60S contains additional masses located near the exit tunnel (red), below the L1-stalk (orange), around the central protuberance (yellow), in the center of the inter-subunit surface (green), next to the stalk base (cyan and blue), in the P-site (cyan), and between below the stalk base and exit tunnel region (purple). The crown view (upper panel) is rotated by 90° as indicated. Arrowheads indicate helix 69. (b) Comparison of the Arx1-particle reconstruction (right) with negative stain EM class averages (left) both affinity-purified via Alb1-TAP at 5 mM MgCl2. Colors as in a. Characteristic pre-ribosomal structures of the Arx1-particle are labeled (foot, orange; nose, yellow and parts of the central protuberance; knob, red). Scale bar, 10 nm. (c) A 60S subunit model containing ES27 in out conformation (ribbon; PDB: 3IZF, 3IZD, 3IZS) was fitted into the Arx1-particle reconstruction. The close-up (right) is displayed at higher contour level than the overview (left). In most areas the model fits well into the density (left). No density was observed for the mature positions of A-site finger (purple) and 5S rRNA (red; right). The density found at the mature location of Rpl10 (orange) does not fit its shape and displays typical features of RNA density. Abbreviations as in a.
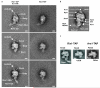
Figure 3. Immuno-EM reveals the relative position of biogenesis factors and r-proteins on the Arx1-particle
(a) Class averages of negative-stained Arx1-particles affinity-purified via Alb1-TAP with anti-HA antibody bound to the indicated C-terminally integrated HA-tagged proteins of interest. The three typical orientations of the Arx1-particle are depicted (view 1–3). The white arrow heads indicate the antibody-derived extra density as compared to control Arx1-particle (Alb1-TAP with integrated Arx1-HA) without added antibody (no anti-HA). Scale bar, 10 nm. (b) Arx1 localizes to the knob structure of the Arx1-particle, as shown by immuno-labeling in a. Ebp1, the slightly smaller human homolog of Arx1, resembles the knob structure of the Arx1-particle in size and shape. The crystal structure of Ebp1 (red ribbon; PDB: 2Q8K) was fitted into the cryo-EM density attributed to Arx1 (red surface). According to this fit, the Arx1 binding pocket would be facing towards the pre-60S subunit, but it would not be occluded. The fit is shown according to the crown (left) and bottom views (right) of the particle. Colors and abbreviations as in Fig. 2a. (c) The additional mass colored in blue matches the ribosomal anti-association factor Tif6 in position, size, and shape. The crystal structure of Tif6 (blue ribbon; PDB: 1G62) fits well into the blue density. Colors as in Fig. 2a.
Figure 4. CRAC analysis confirms the location of Arx1 near the exit tunnel
(a) CRAC analysis on Arx1-HTP and untagged strain. Total number of hits plotted against the relative location along the rDNA sequence. Arx1 binds helix 59 (H59; 22 hits) and helix 63 (15 hits; upper panel). Peaks marked by an asterisk also appear in the negative control (lower panel). (b) Arx1 positioning in a 25S rRNA model (adapted from ref. 6). Nucleotides found in all hits (peak tips in a): dark green, red; nucleotides found in both experiments: green, orange. ES27 on helix 63 comprises three arms (ES27a–c), ES24 is an extension of helix 59. (c) Arx1 binding presented in a 60S model (PDB: 3IZS, 3IZF, 3IZD, displayed using PyMOL). rRNA, gray; r-proteins, light gray; Rpl5, blue; Rpl3, cyan; Rpl26, magenta; Rpl8, yellow. Nucleotides bound by Arx1 colored as in b. ES27 was described in two conformations. The ES27_out_ conformation (middle, right) brings the main cross-links (red, dark green) close together nearby the exit tunnel (ET), contrary to the ES27_in_ conformation (left). (d) A 60S model (PDB: 3O58) and Ebp1 (see Fig. 3b) were fitted into our Arx1-particle reconstruction. The Arx1-density contacts the subunit close to helix 59 (orange nucleotides red in b), Rpl25 (blue) and Rpl35 (green). Rpl19 (yellow) is nearby. Colors as Fig. 2a. (e) Arx1 (red) contacts ES27 (green) in its out conformation. The Arx1-particle (grey) is shown at contour level 0.45 (left; as Fig. 2a) and 0.06 (right; low-pass filtered between 17– 19 Å).
Figure 5. Arx1 localizes to the knob structure of the Rix1-particle, a precursor of the Arx1-particle
(a) Immuno-EM analysis reveals the position of Arx1 on the Rix1-particle. Rix1-TAP was affinity-purified in the presence of an anti-HA antibody from a strain with genomically integrated Arx1-HA (left panel) or with no HA integration (right panel). The Rix1-particles were analyzed by negative stain EM and single particle analysis, and three typical class averages are shown. The main structural features of the Rix1-particle are indicated as “body”, “Rea1 AAA” (AAA+ ATPase ring), “Rea1 tail” and “knob”. The white arrow points to the additional mass attributed to the bound anti-HA antibody and indicative of the position of Arx1 on the particle. Scale bar, 10 nm. (b) Arx1 localizes to the conspicuous knob structure of the Rix1-particle. The position of Arx1 is shown in relation to the known positions of Rpl3, Rsa4, Rpl5, Ipi3, Rix1, and Rea1-MIDAS determined by immuno-EM on the Rix1-particle (adapted to ref. 14). The “body”, “knob”, “Rea1 AAA”, and “Rea1-MIDAS” are indicated as main features of the Rix1-particle. (c) Comparison of Rix1-particle and Arx1-particle class averages. Comparison of Rix1-TAP particles containing the Rea1-tail and tail-less particles obtained after ATP-treatment (+ATP; adapted from Supplementary Fig. 1a, see ref. 14), and particles affinity-purified via Arx1-TAP. In all cases, the typical “knob” is indicated. Scale bar, 10 nm.
Similar articles
- The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae.
Demoinet E, Jacquier A, Lutfalla G, Fromont-Racine M. Demoinet E, et al. RNA. 2007 Sep;13(9):1570-81. doi: 10.1261/rna.585007. Epub 2007 Jul 24. RNA. 2007. PMID: 17652132 Free PMC article. - Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis.
Rosado IV, Dez C, Lebaron S, Caizergues-Ferrer M, Henry Y, de la Cruz J. Rosado IV, et al. Mol Cell Biol. 2007 Feb;27(4):1207-21. doi: 10.1128/MCB.01523-06. Epub 2006 Dec 4. Mol Cell Biol. 2007. PMID: 17145778 Free PMC article. - The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis.
Dong J, Lai R, Jennings JL, Link AJ, Hinnebusch AG. Dong J, et al. Mol Cell Biol. 2005 Nov;25(22):9859-73. doi: 10.1128/MCB.25.22.9859-9873.2005. Mol Cell Biol. 2005. PMID: 16260602 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Topical fluoride as a cause of dental fluorosis in children.
Wong MCM, Zhang R, Luo BW, Glenny AM, Worthington HV, Lo ECM. Wong MCM, et al. Cochrane Database Syst Rev. 2024 Jun 20;6(6):CD007693. doi: 10.1002/14651858.CD007693.pub3. Cochrane Database Syst Rev. 2024. PMID: 38899538 Review.
Cited by
- Eukaryote-specific rRNA expansion segments function in ribosome biogenesis.
Ramesh M, Woolford JL Jr. Ramesh M, et al. RNA. 2016 Aug;22(8):1153-62. doi: 10.1261/rna.056705.116. Epub 2016 Jun 17. RNA. 2016. PMID: 27317789 Free PMC article. - MetAP-like Ebp1 occupies the human ribosomal tunnel exit and recruits flexible rRNA expansion segments.
Wild K, Aleksić M, Lapouge K, Juaire KD, Flemming D, Pfeffer S, Sinning I. Wild K, et al. Nat Commun. 2020 Feb 7;11(1):776. doi: 10.1038/s41467-020-14603-7. Nat Commun. 2020. PMID: 32034140 Free PMC article. - Identification of the binding site of Rlp7 on assembling 60S ribosomal subunits in Saccharomyces cerevisiae.
Dembowski JA, Ramesh M, McManus CJ, Woolford JL Jr. Dembowski JA, et al. RNA. 2013 Dec;19(12):1639-47. doi: 10.1261/rna.041194.113. Epub 2013 Oct 15. RNA. 2013. PMID: 24129494 Free PMC article. - Targeted cross-linking-mass spectrometry determines vicinal interactomes within heterogeneous RNP complexes.
Trahan C, Oeffinger M. Trahan C, et al. Nucleic Acids Res. 2016 Feb 18;44(3):1354-69. doi: 10.1093/nar/gkv1366. Epub 2015 Dec 10. Nucleic Acids Res. 2016. PMID: 26657640 Free PMC article. - Paradigms of ribosome synthesis: Lessons learned from ribosomal proteins.
Gamalinda M, Woolford JL Jr. Gamalinda M, et al. Translation (Austin). 2015 Feb 2;3(1):e975018. doi: 10.4161/21690731.2014.975018. eCollection 2015 Jan-Jun. Translation (Austin). 2015. PMID: 26779413 Free PMC article. Review.
References
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–83. - PubMed
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annual review of cell and developmental biology. 1999;15:607–60. - PubMed
- Zemp I, Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581:2783–93. - PubMed
- Spahn CM, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous