Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes - PubMed (original) (raw)
Review
Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes
Sarah J Goodfellow et al. Subcell Biochem. 2013.
Abstract
RNA Polymerase (Pol) I produces ribosomal (r)RNA, an essential component of the cellular protein synthetic machinery that drives cell growth, underlying many fundamental cellular processes. Extensive research into the mechanisms governing transcription by Pol I has revealed an intricate set of control mechanisms impinging upon rRNA production. Pol I-specific transcription factors guide Pol I to the rDNA promoter and contribute to multiple rounds of transcription initiation, promoter escape, elongation and termination. In addition, many accessory factors are now known to assist at each stage of this transcription cycle, some of which allow the integration of transcriptional activity with metabolic demands. The organisation and accessibility of rDNA chromatin also impinge upon Pol I output, and complex mechanisms ensure the appropriate maintenance of the epigenetic state of the nucleolar genome and its effective transcription by Pol I. The following review presents our current understanding of the components of the Pol I transcription machinery, their functions and regulation by associated factors, and the mechanisms operating to ensure the proper transcription of rDNA chromatin. The importance of such stringent control is demonstrated by the fact that deregulated Pol I transcription is a feature of cancer and other disorders characterised by abnormal translational capacity.
Figures
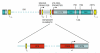
Fig. 10.1. Mammalian rDNA repeat and 47S rRNA promoter
The top panel illustrates key elements and the general organisation of a mammalian rDNA repeat. The IGS includes the spacer and 47S rRNA promoters, enhancer repeats and the TTF-I binding sites T0 and Tsp. Arrows indicate start sites and direction of transcription. The coding region contains 5′ and 3′ external transcribed spacer (ETS) and two internal transcribed spacer (ITS) regions, along with regions encoding 18S, 5.8S and 28S rRNAs. Terminator elements (T1-10) downstream of the 47S rRNA gene are also indicated. The lower panel illustrates the layout of the 47S rRNA promoter, which directs the assembly of the Pol I PIC and consists of an upstream control element, and a core promoter element overlapping the transcription start site.
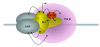
Fig. 10.2. The mammalian Pol I pre-initiation complex
Activated transcription by Pol I requires the assembly of Pol I-specific transcription factors SL1 and UBF at the rRNA promoter. In addition to contacts made between these transcription factors and the rDNA, several protein-protein interactions are also known to facilitate PIC assembly, as indicated by double-headed arrows (described in the text). A multitude of other factors cooperate with this transcription machinery to enhance PIC assembly and promote efficient rRNA synthesis by Pol I in vivo.
Similar articles
- Regulation of RNA Polymerase I Transcription in Development, Disease, and Aging.
Sharifi S, Bierhoff H. Sharifi S, et al. Annu Rev Biochem. 2018 Jun 20;87:51-73. doi: 10.1146/annurev-biochem-062917-012612. Epub 2018 Mar 28. Annu Rev Biochem. 2018. PMID: 29589958 Review. - Yeast transcription elongation factor Spt5 associates with RNA polymerase I and RNA polymerase II directly.
Viktorovskaya OV, Appling FD, Schneider DA. Viktorovskaya OV, et al. J Biol Chem. 2011 May 27;286(21):18825-33. doi: 10.1074/jbc.M110.202119. Epub 2011 Apr 5. J Biol Chem. 2011. PMID: 21467036 Free PMC article. - Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2.
Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Young DW, et al. Nature. 2007 Jan 25;445(7126):442-6. doi: 10.1038/nature05473. Nature. 2007. PMID: 17251981 - Beyond rRNA: nucleolar transcription generates a complex network of RNAs with multiple roles in maintaining cellular homeostasis.
Feng S, Manley JL. Feng S, et al. Genes Dev. 2022 Aug 1;36(15-16):876-886. doi: 10.1101/gad.349969.122. Genes Dev. 2022. PMID: 36207140 Free PMC article. Review. - Hmo1 Promotes Efficient Transcription Elongation by RNA Polymerase I in Saccharomyces cerevisiae.
Huffines AK, Schneider DA. Huffines AK, et al. Genes (Basel). 2024 Feb 15;15(2):247. doi: 10.3390/genes15020247. Genes (Basel). 2024. PMID: 38397236 Free PMC article.
Cited by
- Mammary epithelial morphogenesis and early breast cancer. Evidence of involvement of basal components of the RNA Polymerase I transcription machinery.
Rossetti S, Wierzbicki AJ, Sacchi N. Rossetti S, et al. Cell Cycle. 2016 Sep 16;15(18):2515-26. doi: 10.1080/15384101.2016.1215385. Epub 2016 Aug 2. Cell Cycle. 2016. PMID: 27485818 Free PMC article. - Improvement of ribonucleic acid production in Cyberlindnera jadinii and optimization of fermentation medium.
Li M, Gao S, Yang P, Li H. Li M, et al. AMB Express. 2024 Feb 15;14(1):24. doi: 10.1186/s13568-024-01679-3. AMB Express. 2024. PMID: 38358520 Free PMC article. - RNA Polymerases I and III in development and disease.
Watt KE, Macintosh J, Bernard G, Trainor PA. Watt KE, et al. Semin Cell Dev Biol. 2023 Feb 28;136:49-63. doi: 10.1016/j.semcdb.2022.03.027. Epub 2022 Apr 11. Semin Cell Dev Biol. 2023. PMID: 35422389 Free PMC article. Review. - AF4 uses the SL1 components of RNAP1 machinery to initiate MLL fusion- and AEP-dependent transcription.
Okuda H, Kanai A, Ito S, Matsui H, Yokoyama A. Okuda H, et al. Nat Commun. 2015 Nov 23;6:8869. doi: 10.1038/ncomms9869. Nat Commun. 2015. PMID: 26593443 Free PMC article. - Timing-specific parental effects of ocean warming in a coral reef fish.
Bonzi LC, Spinks RK, Donelson JM, Munday PL, Ravasi T, Schunter C. Bonzi LC, et al. Proc Biol Sci. 2024 May;291(2023):20232207. doi: 10.1098/rspb.2023.2207. Epub 2024 May 22. Proc Biol Sci. 2024. PMID: 38772423 Free PMC article.
References
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG, Wright AP. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. - PubMed
- Ayrault O, Andrique L, Fauvin D, Eymin B, Gazzeri S, Seite P. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene. 2006;25:7577–7586. - PubMed
- Bazett-Jones DP, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources