Analyses of pig genomes provide insight into porcine demography and evolution - PubMed (original) (raw)
. 2012 Nov 15;491(7424):393-8.
doi: 10.1038/nature11622.
Alan L Archibald, Hirohide Uenishi, Christopher K Tuggle, Yasuhiro Takeuchi, Max F Rothschild, Claire Rogel-Gaillard, Chankyu Park, Denis Milan, Hendrik-Jan Megens, Shengting Li, Denis M Larkin, Heebal Kim, Laurent A F Frantz, Mario Caccamo, Hyeonju Ahn, Bronwen L Aken, Anna Anselmo, Christian Anthon, Loretta Auvil, Bouabid Badaoui, Craig W Beattie, Christian Bendixen, Daniel Berman, Frank Blecha, Jonas Blomberg, Lars Bolund, Mirte Bosse, Sara Botti, Zhan Bujie, Megan Bystrom, Boris Capitanu, Denise Carvalho-Silva, Patrick Chardon, Celine Chen, Ryan Cheng, Sang-Haeng Choi, William Chow, Richard C Clark, Christopher Clee, Richard P M A Crooijmans, Harry D Dawson, Patrice Dehais, Fioravante De Sapio, Bert Dibbits, Nizar Drou, Zhi-Qiang Du, Kellye Eversole, João Fadista, Susan Fairley, Thomas Faraut, Geoffrey J Faulkner, Katie E Fowler, Merete Fredholm, Eric Fritz, James G R Gilbert, Elisabetta Giuffra, Jan Gorodkin, Darren K Griffin, Jennifer L Harrow, Alexander Hayward, Kerstin Howe, Zhi-Liang Hu, Sean J Humphray, Toby Hunt, Henrik Hornshøj, Jin-Tae Jeon, Patric Jern, Matthew Jones, Jerzy Jurka, Hiroyuki Kanamori, Ronan Kapetanovic, Jaebum Kim, Jae-Hwan Kim, Kyu-Won Kim, Tae-Hun Kim, Greger Larson, Kyooyeol Lee, Kyung-Tai Lee, Richard Leggett, Harris A Lewin, Yingrui Li, Wansheng Liu, Jane E Loveland, Yao Lu, Joan K Lunney, Jian Ma, Ole Madsen, Katherine Mann, Lucy Matthews, Stuart McLaren, Takeya Morozumi, Michael P Murtaugh, Jitendra Narayan, Dinh Truong Nguyen, Peixiang Ni, Song-Jung Oh, Suneel Onteru, Frank Panitz, Eung-Woo Park, Hong-Seog Park, Geraldine Pascal, Yogesh Paudel, Miguel Perez-Enciso, Ricardo Ramirez-Gonzalez, James M Reecy, Sandra Rodriguez-Zas, Gary A Rohrer, Lauretta Rund, Yongming Sang, Kyle Schachtschneider, Joshua G Schraiber, John Schwartz, Linda Scobie, Carol Scott, Stephen Searle, Bertrand Servin, Bruce R Southey, Goran Sperber, Peter Stadler, Jonathan V Sweedler, Hakim Tafer, Bo Thomsen, Rashmi Wali, Jian Wang, Jun Wang, Simon White, Xun Xu, Martine Yerle, Guojie Zhang, Jianguo Zhang, Jie Zhang, Shuhong Zhao, Jane Rogers, Carol Churcher, Lawrence B Schook
Affiliations
- PMID: 23151582
- PMCID: PMC3566564
- DOI: 10.1038/nature11622
Analyses of pig genomes provide insight into porcine demography and evolution
Martien A M Groenen et al. Nature. 2012.
Abstract
For 10,000 years pigs and humans have shared a close and complex relationship. From domestication to modern breeding practices, humans have shaped the genomes of domestic pigs. Here we present the assembly and analysis of the genome sequence of a female domestic Duroc pig (Sus scrofa) and a comparison with the genomes of wild and domestic pigs from Europe and Asia. Wild pigs emerged in South East Asia and subsequently spread across Eurasia. Our results reveal a deep phylogenetic split between European and Asian wild boars ∼1 million years ago, and a selective sweep analysis indicates selection on genes involved in RNA processing and regulation. Genes associated with immune response and olfaction exhibit fast evolution. Pigs have the largest repertoire of functional olfactory receptor genes, reflecting the importance of smell in this scavenging animal. The pig genome sequence provides an important resource for further improvements of this important livestock species, and our identification of many putative disease-causing variants extends the potential of the pig as a biomedical model.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
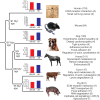
Figure 1. Phylogeny of the six mammals used in the dN/dS analysis.
KEGG pathways with genes that show accelerated evolution for each of the six mammals used in the dN/dS analysis. The bar charts show the individual dN/dS and dS values for each of the six mammals. The dN/dS and dS values refer to the time period of each of the six individual lineages. The number of proteins that show significantly accelerated dN/dS ratios in each lineage varies from 84 in the mouse to 311 in the pig lineage. Pathways significantly (P < 0.05) enriched within this group of genes are also shown with the number of genes shown in brackets. HPI, Helicobacter pylori infection. PowerPoint slide
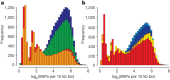
Figure 2. Distribution of heterozygosity for individual pig genomes.
Shown is the distribution of the heterozygosity as the log2(SNPs) per 10k bin. a, Wild Sus scrofa: blue, south China; green, north China; orange, Italian; red, Dutch. b, Breeds: blue, Chinese breeds (Jiangquhai, Meishan, Xiang); red–yellow, European breeds (Hampshire, large white, landrace). Note that the Hampshire breed is a North American breed of European origin. PowerPoint slide
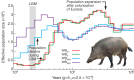
Figure 3. Demographic history of wild boars.
Demographic history was inferred using a hidden Markov model (HMM) approach as implemented in pairwise sequentially Markovian coalescence (PSMC). In the absence of known mutation rates for pig, we used the default mutation rate for human (μ) of 2.5 × 10−8. For the generation time (g) we used an estimate of 5 years. The Last Glacial Maximum (LGM) is highlighted in grey. WBnl, wild boar Netherlands; WBit, wild boar Italy; WBNch, wild boar north China; WBSch, wild boar south China. PowerPoint slide
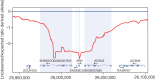
Figure 4. Putative selective sweep region around the ERI2 gene on SSC3.
The y axis shows the log-transformed value of the ratio for the observed/expected derived allele frequency using a sliding window at a bin size of 50,000 bp. The x axis shows the position on SSC3 in base pairs. PowerPoint slide
Similar articles
- A decade of pig genome sequencing: a window on pig domestication and evolution.
Groenen MA. Groenen MA. Genet Sel Evol. 2016 Mar 29;48:23. doi: 10.1186/s12711-016-0204-2. Genet Sel Evol. 2016. PMID: 27025270 Free PMC article. Review. - Molecular microevolution and epigenetic patterns of the long non-coding gene H19 show its potential function in pig domestication and breed divergence.
Li C, Wang X, Cai H, Fu Y, Luan Y, Wang W, Xiang H, Li C. Li C, et al. BMC Evol Biol. 2016 Apr 23;16:87. doi: 10.1186/s12862-016-0657-5. BMC Evol Biol. 2016. PMID: 27107967 Free PMC article. - Insular East Asia pig dispersal and vicariance inferred from Asian wild boar genetic evidence.
Li KY, Li KT, Yang CH, Hwang MH, Chang SW, Lin SM, Wu HJ, Basilio EB, Vega RSA, Laude RP, Ju YT. Li KY, et al. J Anim Sci. 2017 Apr;95(4):1451-1466. doi: 10.2527/jas.2016.1117. J Anim Sci. 2017. PMID: 28464072 - Genome-wide SNP data unveils the globalization of domesticated pigs.
Yang B, Cui L, Perez-Enciso M, Traspov A, Crooijmans RPMA, Zinovieva N, Schook LB, Archibald A, Gatphayak K, Knorr C, Triantafyllidis A, Alexandri P, Semiadi G, Hanotte O, Dias D, Dovč P, Uimari P, Iacolina L, Scandura M, Groenen MAM, Huang L, Megens HJ. Yang B, et al. Genet Sel Evol. 2017 Sep 21;49(1):71. doi: 10.1186/s12711-017-0345-y. Genet Sel Evol. 2017. PMID: 28934946 Free PMC article. - Mining the pig genome to investigate the domestication process.
Ramos-Onsins SE, Burgos-Paz W, Manunza A, Amills M. Ramos-Onsins SE, et al. Heredity (Edinb). 2014 Dec;113(6):471-84. doi: 10.1038/hdy.2014.68. Epub 2014 Jul 30. Heredity (Edinb). 2014. PMID: 25074569 Free PMC article. Review.
Cited by
- Copy Number Variation and Selection Signal: Exploring the Domestication History and Phenotype Differences Between Duroc and the Chinese Native Ningxiang Pigs.
Yang F, Chen W, Yang Y, Meng Y, Chen Y, Ding X, Zhang Y, He J, Gao N. Yang F, et al. Int J Mol Sci. 2024 Oct 31;25(21):11716. doi: 10.3390/ijms252111716. Int J Mol Sci. 2024. PMID: 39519268 Free PMC article. - Genetic Adaptations of the Tibetan Pig to High-Altitude Hypoxia on the Qinghai-Tibet Plateau.
Yang Y, Yuan H, Yao B, Zhao S, Wang X, Xu L, Zhang L. Yang Y, et al. Int J Mol Sci. 2024 Oct 21;25(20):11303. doi: 10.3390/ijms252011303. Int J Mol Sci. 2024. PMID: 39457085 Free PMC article. - Polymorphism Identification in the Coding Sequences (ORFs) of the Porcine Pregnancy-Associated Glycoprotein 2-like Gene Subfamily in Pigs.
Bieniek-Kobuszewska M, Panasiewicz G. Bieniek-Kobuszewska M, et al. Genes (Basel). 2024 Aug 31;15(9):1149. doi: 10.3390/genes15091149. Genes (Basel). 2024. PMID: 39336740 Free PMC article. - A genome assembly and transcriptome atlas of the inbred Babraham pig to illuminate porcine immunogenetic variation.
Schwartz JC, Farrell CP, Freimanis G, Sewell AK, Phillips JD, Hammond JA. Schwartz JC, et al. Immunogenetics. 2024 Dec;76(5-6):361-380. doi: 10.1007/s00251-024-01355-7. Epub 2024 Sep 19. Immunogenetics. 2024. PMID: 39294478 Free PMC article. - A comprehensive atlas of nuclear sequences of mitochondrial origin (NUMT) inserted into the pig genome.
Bolner M, Bovo S, Ballan M, Schiavo G, Taurisano V, Ribani A, Bertolini F, Fontanesi L. Bolner M, et al. Genet Sel Evol. 2024 Sep 16;56(1):64. doi: 10.1186/s12711-024-00930-6. Genet Sel Evol. 2024. PMID: 39285356 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- T32 AI083196/AI/NIAID NIH HHS/United States
- 5 P41 LM006252/LM/NLM NIH HHS/United States
- R13 RR032267/RR/NCRR NIH HHS/United States
- R21 DA027548/DA/NIDA NIH HHS/United States
- BBS/E/D/20211550/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E010520/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 095908/WT_/Wellcome Trust/United Kingdom
- 249894/ERC_/European Research Council/International
- R13 RR020283A/RR/NCRR NIH HHS/United States
- BB/E010768/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- P41 LM006252/LM/NLM NIH HHS/United States
- BB/E010520/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0900950/MRC_/Medical Research Council/United Kingdom
- P20 RR017686/RR/NCRR NIH HHS/United States
- BB/H005935/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I025328/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R21 HG006464/HG/NHGRI NIH HHS/United States
- P30 DA018310/DA/NIDA NIH HHS/United States
- R13 RR032267A/RR/NCRR NIH HHS/United States
- R13 RR020283/RR/NCRR NIH HHS/United States
- 5 P41LM006252/LM/NLM NIH HHS/United States
- ETM/32/CSO_/Chief Scientist Office/United Kingdom
- BB/G004013/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/05191130/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- P20-RR017686/RR/NCRR NIH HHS/United States
- BB/E011640/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources