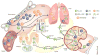Re(de)fining the dendritic cell lineage - PubMed (original) (raw)
Review
. 2012 Dec;13(12):1145-54.
doi: 10.1038/ni.2467. Epub 2012 Nov 16.
Affiliations
- PMID: 23160217
- PMCID: PMC3644874
- DOI: 10.1038/ni.2467
Review
Re(de)fining the dendritic cell lineage
Ansuman T Satpathy et al. Nat Immunol. 2012 Dec.
Erratum in
- Nat Immunol. 2013 Apr;14(4):413
Abstract
Dendritic cells (DCs) are essential mediators of innate and adaptive immune responses. Study of these critical cells has been complicated by their similarity to other hematopoietic lineages, particularly monocytes and macrophages. Progress has been made in three critical areas of DC biology: the characterization of lineage-restricted progenitors in the bone marrow, the identification of cytokines and transcription factors required during differentiation, and the development of genetic tools for the visualization and depletion of DCs in vivo. Collectively, these advances have clarified the nature of the DC lineage and have provided novel insights into their function during health and disease.
Conflict of interest statement
The authors have no conflicting financial interests.
Figures
Figure 1. Development and migration of mononuclear phagocyte lineages in the steady state
Classical DCs (cDCs), plasmacytoid DCs (pDCs) and monocytes (Mono) derive from bone marrow (BM) progenitors. Macrophage-DC progenitors (MDPs) give rise to common dendritic cell progenitors (CDPs) and monocytes. CDPs differentiate into pDCs or committed precursors for cDCs (pre-cDCs). Pre-cDCs, pDCs, and monocytes transit through the blood and seed peripheral organs, where pre-cDCs complete their differentiation into CD8+/CD103+ or CD4+/CD11b+ cDCs. Monocytes can migrate into tissues and differentiate into macrophages. In the intestine, cDCs and macrophages populate the villi; cDCs are also present in intestinal lymphoid follicles (ILFs). In the skin, dermal DCs consist of both CD11b+ and CD103+ cDC subsets. Langerhans cells (LCs) populate the epidermis and self-renew locally. Macrophages, pDCs and both cDC subsets reside in the lung. A hallmark characteristic of cDCs is their ability to migrate upon antigen encounter from tissues to draining lymph nodes to prime T cell responses. In contrast, macrophages largely remain at the site of differentiation.
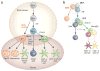
Figure 2. Global relationships between DC and macrophage subsets
Principal component analysis (PCA) of mature macrophage and DC subsets is shown. Each circle represents at least two microarray replicates. (a,b) Samples segregate by organ (PC1 and PC2) and by lineage (PC3). Genes with the greatest positive and negative loadings in PC3 reflect their DC- or macrophage-specific expression, respectively. Values in parentheses indicate proportion of variance explained. (c) Gene expression levels of Mafb and Zbtb46 in cDC and macrophage subsets are compared against PC3 scores from (b). MF, macrophage; PC, principal component; RPM, red pulp macrophage; SI, small intestine.
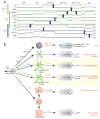
Figure 3. Distinguishing myeloid populations with lineage-specific transcription factors
(a) Lineage-specific transcription factor expression represents an alternative to surface marker-based methods for accurately identifying myeloid cell types. Lineage- or stage-specific transcription factors are indicated in colors corresponding to cell types in which they are uniquely expressed. For example, E2-2 expression distinguishes the pDC lineage while Zbtb46 distinguishes cDCs. (b) Shown is the theoretical heterogeneity within progenitor stages as defined by FACS (dashed circles). For example, cells characterized as CDPs using cell surface markers include a subpopulation of cells already committed to the cDC lineage, which are identified by expression of Zbtb46.
Figure 4. Stage-specific expression of transcription factors controlling DC development and specialization
(a) Shown are approximate mRNA expression levels,, of selected transcription factors regulating DC development. Factors are grouped by the lineage in which they are required. Vertical bars indicate the stage at which each factor is essential for development. (b) Shown are sequential transcription factor requirements during development. Analysis of models deficient in these factors suggests that an important consequence of subset specialization is the ability to respond differentially to pathogen challenge through the secretion of specific cytokines. This specificity closely resembles the cytokine requirements for CD4+ T helper (TH) subset differentiation, highlighting that DC responses are a key determinant of the resulting adaptive immune response. As in T cells, DCs may be better characterized by function rather than surface marker expression, i.e. CD8+ cDCs as “DC12” cells.
Similar articles
- Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages.
Satpathy AT, KC W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, Murphy TL, Murphy KM. Satpathy AT, et al. J Exp Med. 2012 Jun 4;209(6):1135-52. doi: 10.1084/jem.20120030. Epub 2012 May 21. J Exp Med. 2012. PMID: 22615127 Free PMC article. - Dendritic cell subsets and lineages, and their functions in innate and adaptive immunity.
Liu YJ. Liu YJ. Cell. 2001 Aug 10;106(3):259-62. doi: 10.1016/s0092-8674(01)00456-1. Cell. 2001. PMID: 11509173 Review. No abstract available. - A common pathway for dendritic cell and early B cell development.
Izon D, Rudd K, DeMuth W, Pear WS, Clendenin C, Lindsley RC, Allman D. Izon D, et al. J Immunol. 2001 Aug 1;167(3):1387-92. doi: 10.4049/jimmunol.167.3.1387. J Immunol. 2001. PMID: 11466357 - Epigenetic control of Ccr7 expression in distinct lineages of lung dendritic cells.
Moran TP, Nakano H, Kondilis-Mangum HD, Wade PA, Cook DN. Moran TP, et al. J Immunol. 2014 Nov 15;193(10):4904-13. doi: 10.4049/jimmunol.1401104. Epub 2014 Oct 8. J Immunol. 2014. PMID: 25297875 Free PMC article. - Transcription factor networks in dendritic cell development.
Satpathy AT, Murphy KM, KC W. Satpathy AT, et al. Semin Immunol. 2011 Oct;23(5):388-97. doi: 10.1016/j.smim.2011.08.009. Epub 2011 Sep 15. Semin Immunol. 2011. PMID: 21924924 Free PMC article. Review.
Cited by
- The interaction of innate immune and adaptive immune system.
Wang R, Lan C, Benlagha K, Camara NOS, Miller H, Kubo M, Heegaard S, Lee P, Yang L, Forsman H, Li X, Zhai Z, Liu C. Wang R, et al. MedComm (2020). 2024 Sep 15;5(10):e714. doi: 10.1002/mco2.714. eCollection 2024 Oct. MedComm (2020). 2024. PMID: 39286776 Free PMC article. Review. - The Multifaceted Functionality of Plasmacytoid Dendritic Cells in Gastrointestinal Cancers: A Potential Therapeutic Target?
Hansen FJ, David P, Weber GF. Hansen FJ, et al. Cancers (Basel). 2024 Jun 13;16(12):2216. doi: 10.3390/cancers16122216. Cancers (Basel). 2024. PMID: 38927922 Free PMC article. Review. - Dendritic Cells in Shaping Anti-Tumor T Cell Response.
Mazzoccoli L, Liu B. Mazzoccoli L, et al. Cancers (Basel). 2024 Jun 13;16(12):2211. doi: 10.3390/cancers16122211. Cancers (Basel). 2024. PMID: 38927916 Free PMC article. Review. - Differential Response of Human Dendritic Cells upon Stimulation with Encapsulated or Non-Encapsulated Isogenic Strains of Porphyromonas gingivalis.
Melgar-Rodríguez S, Polanco A, Ríos-Muñoz J, García M, Sierra-Cristancho A, González-Osuna L, Díaz-Zúñiga J, Carvajal P, Vernal R, Bravo D. Melgar-Rodríguez S, et al. Int J Mol Sci. 2024 Apr 20;25(8):4510. doi: 10.3390/ijms25084510. Int J Mol Sci. 2024. PMID: 38674095 Free PMC article. - The role of dendritic cells in MASH: friends or foes?
Pinto AT, Lukacs-Kornek V. Pinto AT, et al. Front Immunol. 2024 Apr 8;15:1379225. doi: 10.3389/fimmu.2024.1379225. eCollection 2024. Front Immunol. 2024. PMID: 38650949 Free PMC article. Review.
References
- Xie H, et al. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. - PubMed
- Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. - PubMed
- Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. - PubMed
- Steinman RM. Decisions About Dendritic Cells: Past, Present, and Future. Annu Rev Immunol. 2011;30:1–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- P30 CA91842/CA/NCI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- AI076427-02/AI/NIAID NIH HHS/United States
- R01 AI076427/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources