Simulating realistic genomic data with rare variants - PubMed (original) (raw)
Simulating realistic genomic data with rare variants
Yaji Xu et al. Genet Epidemiol. 2013 Feb.
Abstract
Increasing evidence suggests that rare and generally deleterious genetic variants might have a strong impact on disease risks of not only Mendelian disease, but also many common diseases. However, identifying such rare variants remains challenging, and novel statistical methods and bioinformatic software must be developed. Hence, we have to extensively evaluate various methods under reasonable genetic models. Although there are abundant genomic data, they are not most helpful for the evaluation of the methods because the disease mechanism is unknown. Thus, it is imperative that we simulate genomic data that mimic the real data containing rare variants and that enable us to impose a known disease penetrance model. Although resampling simulation methods have shown their advantages in computational efficiency and in preserving important properties such as linkage disequilibrium (LD) and allele frequency, they still have limitations as we demonstrated. We propose an algorithm that combines a regression-based imputation with resampling to simulate genetic data with both rare and common variants. Logistic regression model was employed to fit the relationship between a rare variant and its nearby common variants in the 1000 Genomes Project data and then applied to the real data to fill in one rare variant at a time using the fitted logistic model based on common variants. Individuals then were simulated using the real data with imputed rare variants. We compared our method with existing simulators and demonstrated that our method performed well in retaining the real sample properties, such as LD and minor allele frequency, qualitatively.
© 2012 WILEY PERIODICALS, INC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
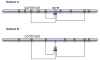
Figure 1. Simulating rare variants using datasets A and B
For a rare variant, a logistic regression model was built in dataset A using selected common variants surrounding, and this rare variant is then imputed in dataset B based on the constructed logistic model.
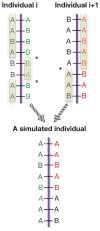
Figure 2. Simulating a genotype vector from two individuals (four haplotype vectors)
Started with the genomic data of individuals i and i + 1, a new individual is simulated by crossing over a pair of haplotypes from individual i, and a pair of haplotypes from individual i + 1. An asterisk * in the figure indicates the location where a cross-over event occurs. The alleles in the rectangles are selected to form the new individual.
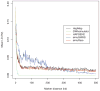
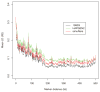
Figure 3. LD patterns by different simulation methods and their reference (HapMap CEU)
Each curve demonstrates the change of the averaged value over pairwise LD (_R_2) values along different marker distances for four simulators, and their reference HapMap CEU sample (black). Note that simuRare implements our regression-based resampling approach.
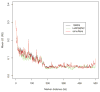
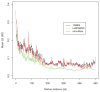
Figure 4. LD patterns by different simulation methods and their reference (1000 Genomes)
Each curve demonstrates the change of the averaged value over pairwise LD (_R_2) values along different marker distances for HAPGEN2 and regression-based algorithm, and their reference the 1000 Genomes Phase I interim sample (black).
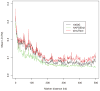
Figure 5. LD patterns by different simulation methods and their reference (1000 Genomes without rare SNPs)
Each curve demonstrates the change of the averaged value over pairwise LD (_R_2) values along different marker distances for HAPGEN2 and regression-based algorithm, and their reference 1000 Genomes Phase I interim sample after removing SNPs with MAF less than 0.01 (black).
Figure 6. LD patterns by different simulation methods and their reference (1000 Genomes CEU)
Each curve demonstrates the change of the averaged value over pairwise LD (_R_2) values along different marker distances for HAPGEN2 and regression-based algorithm, and their reference the 1000 Genomes Phase I interim CEU sample (black).
Figure 7. LD patterns by different simulation methods and their reference (1000 Genomes CEU without rare SNPs)
Each curve demonstrates the change of the averaged value over pairwise LD (_R_2) values along different marker distances for HAPGEN2 and regression-based algorithm, and their reference the 1000 Genomes Phase I interim CEU sample after removing SNPs with MAF less than 0.01 (black).
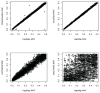
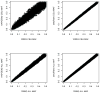
Figure 8. Simulated allele frequencies from four simulators against the reference (HapMap CEU)
Each panel in this figure demonstrates the comparison of the MAFs from one simulator with those from the reference HapMap CEU sample with imputed rare SNPs. Each point represents the deviation of a simulated allele frequency from the real allele frequency obtained from HapMap CEU sample.
Figure 9. Simulated allele frequencies from two simulators against the reference (1000 Genomes)
Each panel in this figure demonstrates the comparison of the MAFs from one simulator with those from the reference 1000 Genomes CEU or mixed population with all individuals from Africa, Asia, Europe, and the Americas. Each point represents the deviation of a simulated allele frequency from the real allele frequency obtained from the 1000 Genomes CEU or mixed sample.
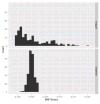
Figure 10. Histograms of the simulated MAF errors for the 1000 Genomes CEU sample
The upper panel is the plot for HAPGEN2, and the lower panel is the plot for simuRare.
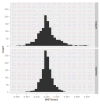
Figure 11. Histograms of the simulated MAF errors for the 1000 Genomes mixed sample
The upper panel is the plot for HAPGEN2, and the lower panel is the plot for simuRare.
Similar articles
- sim1000G: a user-friendly genetic variant simulator in R for unrelated individuals and family-based designs.
Dimitromanolakis A, Xu J, Krol A, Briollais L. Dimitromanolakis A, et al. BMC Bioinformatics. 2019 Jan 15;20(1):26. doi: 10.1186/s12859-019-2611-1. BMC Bioinformatics. 2019. PMID: 30646839 Free PMC article. - Joint association testing of common and rare genetic variants using hierarchical modeling.
Cardin NJ, Mefford JA, Witte JS. Cardin NJ, et al. Genet Epidemiol. 2012 Sep;36(6):642-51. doi: 10.1002/gepi.21659. Epub 2012 Jul 16. Genet Epidemiol. 2012. PMID: 22807252 Free PMC article. - Are rare variants really independent?
Turkmen A, Lin S. Turkmen A, et al. Genet Epidemiol. 2017 May;41(4):363-371. doi: 10.1002/gepi.22039. Epub 2017 Mar 16. Genet Epidemiol. 2017. PMID: 28300291 - Simulating linkage disequilibrium structures in a human population for SNP association studies.
Yuan X, Zhang J, Wang Y. Yuan X, et al. Biochem Genet. 2011 Jun;49(5-6):395-409. doi: 10.1007/s10528-011-9416-x. Epub 2011 Jan 14. Biochem Genet. 2011. PMID: 21234669 Free PMC article. - Simulating sequences of the human genome with rare variants.
Peng B, Liu X. Peng B, et al. Hum Hered. 2010;70(4):287-91. doi: 10.1159/000323316. Epub 2011 Jan 6. Hum Hered. 2010. PMID: 21212684 Free PMC article.
Cited by
- Second-generation PLINK: rising to the challenge of larger and richer datasets.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Chang CC, et al. Gigascience. 2015 Feb 25;4:7. doi: 10.1186/s13742-015-0047-8. eCollection 2015. Gigascience. 2015. PMID: 25722852 Free PMC article. - PGsim: A Comprehensive and Highly Customizable Personal Genome Simulator.
Juan L, Wang Y, Jiang J, Yang Q, Jiang Q, Wang Y. Juan L, et al. Front Bioeng Biotechnol. 2020 Jan 28;8:28. doi: 10.3389/fbioe.2020.00028. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32047747 Free PMC article. - TARV: tree-based analysis of rare variants identifying risk modifying variants in CTNNA2 and CNTNAP2 for alcohol addiction.
Song C, Zhang H. Song C, et al. Genet Epidemiol. 2014 Sep;38(6):552-9. doi: 10.1002/gepi.21843. Epub 2014 Jul 15. Genet Epidemiol. 2014. PMID: 25041903 Free PMC article. - sim1000G: a user-friendly genetic variant simulator in R for unrelated individuals and family-based designs.
Dimitromanolakis A, Xu J, Krol A, Briollais L. Dimitromanolakis A, et al. BMC Bioinformatics. 2019 Jan 15;20(1):26. doi: 10.1186/s12859-019-2611-1. BMC Bioinformatics. 2019. PMID: 30646839 Free PMC article. - Reproducible simulations of realistic samples for next-generation sequencing studies using Variant Simulation Tools.
Peng B. Peng B. Genet Epidemiol. 2015 Jan;39(1):45-52. doi: 10.1002/gepi.21867. Epub 2014 Nov 13. Genet Epidemiol. 2015. PMID: 25395236 Free PMC article.
References
- Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11(6):415–425. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials