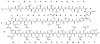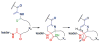Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature - PubMed (original) (raw)
Review
doi: 10.1039/c2np20085f.
Mervyn J Bibb, Gabriele Bierbaum, Albert A Bowers, Tim S Bugni, Grzegorz Bulaj, Julio A Camarero, Dominic J Campopiano, Gregory L Challis, Jon Clardy, Paul D Cotter, David J Craik, Michael Dawson, Elke Dittmann, Stefano Donadio, Pieter C Dorrestein, Karl-Dieter Entian, Michael A Fischbach, John S Garavelli, Ulf Göransson, Christian W Gruber, Daniel H Haft, Thomas K Hemscheidt, Christian Hertweck, Colin Hill, Alexander R Horswill, Marcel Jaspars, Wendy L Kelly, Judith P Klinman, Oscar P Kuipers, A James Link, Wen Liu, Mohamed A Marahiel, Douglas A Mitchell, Gert N Moll, Bradley S Moore, Rolf Müller, Satish K Nair, Ingolf F Nes, Gillian E Norris, Baldomero M Olivera, Hiroyasu Onaka, Mark L Patchett, Joern Piel, Martin J T Reaney, Sylvie Rebuffat, R Paul Ross, Hans-Georg Sahl, Eric W Schmidt, Michael E Selsted, Konstantin Severinov, Ben Shen, Kaarina Sivonen, Leif Smith, Torsten Stein, Roderich D Süssmuth, John R Tagg, Gong-Li Tang, Andrew W Truman, John C Vederas, Christopher T Walsh, Jonathan D Walton, Silke C Wenzel, Joanne M Willey, Wilfred A van der Donk
Affiliations
- PMID: 23165928
- PMCID: PMC3954855
- DOI: 10.1039/c2np20085f
Review
Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature
Paul G Arnison et al. Nat Prod Rep. 2013 Jan.
Abstract
This review presents recommended nomenclature for the biosynthesis of ribosomally synthesized and post-translationally modified peptides (RiPPs), a rapidly growing class of natural products. The current knowledge regarding the biosynthesis of the >20 distinct compound classes is also reviewed, and commonalities are discussed.
Figures
Fig. 1
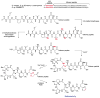
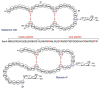
General biosynthetic pathway for RiPPs. The precursor peptide contains a core region that is transformed into the mature product. Many of the post-translational modifications are guided by leader peptide and the recognition sequences as discussed in this review for subclasses of RiPPs. Products of eukaryotic origin also often contain an N-terminal signal peptide that directs the peptide to specialized compartments for modification and secretion. C-terminal recognition sequences are sometimes also present for peptide cyclization (e.g. see sections on cyanobactins, amatoxins, cyclotides, and orbitides). In the case of the bottromycins, a leader-like peptide is appended at the C-terminus of the core peptide.–
Fig. 2
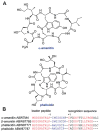
A. Structure of nisin A. Dha residues are shown in green, Dhb residues in purple. Segments of Lan and MeLan originating from Ser/Thr are depicted in red and segments of (Me)Lan originating from Cys in blue. B. Biosynthetic pathway to (Me)Lan and labionin formation. Ser and Thr residues are first phosphorylated and the phosphate is subsequently eliminated to generate Dha and Dhb residues, respectively. Phosphorylation prior to elimination has been experimentally confirmed for class II–IV lanthipeptides and is postulated for class I lanthipeptides. Subsequent intramolecular conjugate addition by the side chain of Cys generates an enolate. Protonation of the enolate produces the (Me)Lan structures whereas attack of the enolate onto another Dha generates the labionin crosslinks.
Fig. 3
Four different classes of lanthipeptides are defined by the four different types of biosynthetic enzymes that install the characteristic thioether crosslinks. The dark areas show conserved regions that are important for catalytic activity. The cyclase domain of class III enzymes has homology with the other cyclase domains/enzymes but lacks the three zinc ligands.
Fig. 4
Some representative lanthipeptides. The same shorthand notation is used as in Fig. 2. Segments of crosslinks originating from Cys are in blue, segments originating from Ser/Thr are in red. Additional post-translational modifications are shown in orange. OBu, 2-oxobutyric acid; Asp-OH, (3_R_)-hydroxy-aspartate; Lys-NH-Ala, lysinoalanine.
Fig. 5
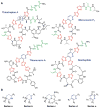
A. Structure of cypemycin, a representative member of the linaridins. The linear sequence of the precursor peptide is shown below the retrosynthetic arrow to illustrate the biosynthetic origin of each PTM: _allo_-Ile from Ile, Dhb from Thr, and AviCys from two Cys residues (underlined). For comparison, the structure of the lanthipeptide epidermin, which also contains an AviCys structure, is shown but its AviCys is formed from a Ser and Cys residue (underlined). B. Chemical structure of AviCys as well as the shorthand notation used in panel A; the absolute configuration of the AviCys and _allo_-Ile in cypemycin is not known.
Fig. 6
Structure of polytheonamide A and B and sequence of the precursor core region. The two congeners differ by the configuration of the sulfoxide moiety. Sulfur oxidation is an artifact occurring during isolation. Color code for posttranslations modifications: purple, epimerization; green, dehydration; red, methylation; blue, hydroxylation.
Fig. 7
A. Proposed biosynthesis of the _N_-acyl unit of polytheonamides. The dehydration step was experimentally verified. B. Polytheonamide biosynthetic gene cluster.
Fig. 8
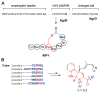
LAP biosynthetic scheme and precursor peptide sequences. A, Upon recognition of the precursor peptide (black line) by the heterocycle-forming BCD enzymatic complex, select Cys and Ser/Thr undergo a backbone cyclodehydration reaction with a preceding amino acid. Catalyzed by a cyclodehydratase (C/D-proteins, green and blue), net loss of water from the amide carbonyl during this reaction yields an azoline heterocycle. Further processing by a FMN-dependent dehydrogenase (B-protein, yellow) affords the aromatic azole heterocycle. B, Amino acid sequence of several LAP precursors. Like most RiPPs, these consist of a leader peptide (to facilitate recognition by the modifying enzymes) and core region that encodes for the resultant natural product. From top to bottom are the LAP precursors for SLS, microcin B17, goadsporin, and plantazolicin. Post-translational modifications are color-coded (Cys resulting in thiazoles, red; Ser/Thr resulting in oxazoles and methyloxazoles, blue; Thr leading to methyloxazoline, brown; Ser converted to dehydroalanine, light blue; N-terminal acetylated/methylated residues, green). The leader peptide cleavage site is indicated with a dash (predicted for SLS).
Fig. 9
Representative structures of LAPs. Microcin B17 is a DNA gyrase inhibitor (antibiotic), goadsporin induces secondary metabolism and morphogenesis in actinomycetes, and plantazolicin is a selective antibiotic with an unknown biological target. Color coding as in Fig. 8.
Fig. 10
Representative cyanobactins. Anacyclamide is modified only by N–C cyclization, while others are modified by heterocyclization of Cys (yellow) or Ser/Thr (blue), oxidation to azole (red dashed line), prenylation (pink) or _N_-methylation (green).
Fig. 11
Generalized biosynthesis of cyanobactins that contain azoline heterocycles. Translation of the core peptide is followed by heterocyclization, then proteolytic tailoring to yield macrocycles. RS = recognition sequence, essential for proteolytic tailoring. Multiple copies of core peptide + recognition sequences are possible; here an example is shown in which a single core peptide exists and is modified (n = 0). Other tailoring, such as prenylation, probably takes place on the mature cycle, while the timing of oxidation/dehydrogenation to azoles is not known. Additional modifications are also sometimes present. Oxi = thiazoline oxidase, N-pro and C-pro are the protease domains that cleave the N-terminus and C-terminus of the core peptides, respectively, and DUF are domains of unknown function. C-pro also performs macrocyclization (Cyc).
Fig. 12
A. Representative thiopeptide structures. The central dehydropiperidine or pyridine rings are in blue, the thiazole and thiazoline rings are in red, and the Dha and Dhb residues are in green. B. The structural series of thiopeptides, showing the typical substitution pattern of the central nitrogenous heterocycle.
Fig. 13
Bycroft proposal for generation of the pyridine ring in thiopeptides from two Dha residues. The aromatization step would also remove the leader peptide.
Fig. 14
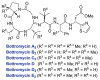
Structures of various bottromycins.
Fig. 15
Potential biosynthetic pathway to bottromycin A2. The order of the posttranslational modifications is currently not known and hence the order depicted is arbitrary. The precursor peptide sequences for the currently known bottromycins are shown at the top. Variations in the posttranslational modifications morph these precursor sequences into the structures shown in Fig. 14.
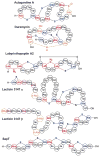
Fig. 16. Structures of the post-translationally modified microcins
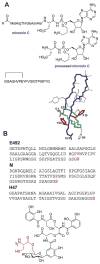
A. Structures of several class I microcins. Both the linear sequence and three dimensional structure of microcin J25 is shown (PDB 1Q71). Microcin B17, another class I microcin, is shown in Fig. 9. B. Class II microcins. The post-translational modifications of class IIb microcins attach a linear trimer of 2,3-dihydroxybenzoyl L-serine linked via a _C_-glycosidic linkage to a β-D-glucose. The glucose is anchored to the carboxylate of the _C_-terminal serine residues of the peptides (red font) through an ester linkage to O6.
Fig. 17. Structural features of lasso peptides
A) Sequences of lasso peptides of the three classes showing the residues involved in the macrolactam in green and those forming the tail in blue. The isopeptide linkage between the N-terminal amino group and the side chain carboxylates of Asp and Glu are shown in dark green. B) Three-dimensional structures of lasso peptides of the three classes; the structures of the class II microcin J25 and capistruin exhibit the respective sizes of the loop and tail above and below the ring.
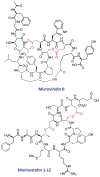
Fig. 18. Structures of microviridin B from M. aeruginosa NIES298 and mar-inostatin 1-12 from Alteromonas sp. B-10-31. ,
Amino acid side chains forming ester and amide bonds are highlighted in red and blue, respectively.
Fig. 19
Sequences of sactipeptides based on structural genes. The arrow indicates the site of cleavage of the N-terminal leader peptide. Subtilosin A and SKF are then cyclized via an amide bond between the amino group of residue 1 and the C-terminal carboxyl. The other peptides are not cyclized in this fashion, but all have cysteine residues linked to α-carbons.
Fig. 20
Three dimensional solution structures of sactipeptides. Subtilosin A has S to Cα crosslinks between C4 and F31, C7 and T28, and C13 and F22. Thurincin H has such links between C4 and S28, C7 and T25, C10 and T22, and C13 and N19. Trn-α has links between C5 and T28, C9 and T25, and C13 and S21. Trn-β has links between C5 and Y28, C9 and A25, and C13 and T21. SKF (not shown, 3D structure not reported) has a single S to Cα crosslink between C4 and M12. It also has a disulfide bond between C1 and C16.
Fig. 21
Comparison of the sequences of some cyclic bacteriocins based on the structural genes. The arrow indicates the cleavage site for the N-terminal leader, whose removal is followed by amide bond formation between the amino group of residue 1 and the carboxyl of the C-terminal residue.
Fig. 22
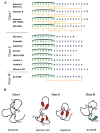
Comparison of the three dimensional structures of enterocin AS-48 (blue) and carnocyclin A (magenta). (A) Sequence comparison with helices and loop indicated. (B) Overlay of backbones of the two peptides showing the saposin fold and similarities in structure. (C) Ribbon diagrams for the two peptides.
Fig. 23
Comparison of gene clusters for biosynthesis of large ring bacteriocins.
Fig. 24
A. Chemical structures of a representative amatoxin and phallotoxin. B. Alignment of a select number of precursor peptides illustrating the highly conserved leader peptide and recognition sequences in red. The core peptide that is post-translationally modified, excised, and cyclized is shown in blue font. For beta-amanitin and phalloidin, the sequences are from genomic DNA whereas the other two sequences are from cDNA. Because of an intron in the genomic DNA, the last three amino acids of the recognition sequence are not known with certainty for beta-amanitin and phalloidin and are represented with dashes.
Fig. 25. Structure of kalata B1 and cyclotide precursor proteins
A schematic representation of three precursor proteins from O. affinis is shown at the top of the figure. The proteins comprise an endoplasmic reticulum signal sequence labeled ER, a leader sequence (comprising a pro-region and a conserved repeated fragment labeled NTR) and either one or multiple copies of the core peptides. A short hydrophobic C-terminal recognition sequence is present in each precursor. At the bottom of the figure the amino acid sequence of kalata B1 is shown, with the cysteine residues labeled with Roman numerals. The cleavage sites for excision of a core cyclotide domain with an N-terminal Gly and a C-terminal Asn, which are thought to be subsequently linked by an asparaginyl endopeptidase, are indicated by arrows. The location of the ligation point, which forms loop 6 of the mature cyclic peptide, is shown on the right. Parts of the precursor protein flanking the mature domain are shown in lighter shading. Figure reprinted with permission from Daly et al.
Fig. 26
Structures of segetalins containing six and seven amino acids, isolated from Vaccaria segetalis (Caryophyllaceae).
Fig. 27
Amino acid sequences of cyclic peptides arising from Linaceae species Linum usitatissimum. The RiPP core peptides putatively encoding cyclolinopeptides D, F, and G is shown in blue, green, and red, respectively, with putative recognition sequences shown in pink and underlined font.
Fig. 28
Structures of three post-translationally modified conopeptides that reached human clinical trials.
Fig. 29. Structures of two glycocins
The precursor peptide that is transformed into sublancin by disulfide formation and _S_-glycosylation is shown below the sublancin 168 structure.
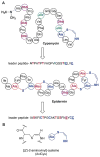
Fig. 30
A. Structure of AIP-I and the ribosomally synthesized precursor peptide from which it is produced. B. Structures of ComX and its derivatives and the ribosomally synthesized precursor peptides from which they are produced. The red font Trp is converted to the structure shown by cyclization of the amide nitrogen of Trp onto C2 of the indole and prenylation at C3. After leader peptide removal, the modified Trp is present in short peptides with the sequences of R1 and R2 shown in blue font.
Fig. 31
Currently accepted structure of methanobactin from M. trichosporium OB3b and the precursor peptide from which it may be derived.
Fig. 32
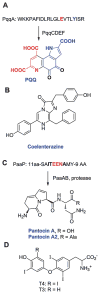
Structures of (A) PQQ, (B) coelenterazine, (C) the pantocins, and (D) the thyroid hormones T3 and T4. For PQQ and the pantocins, the ribosomally synthesized precursor peptides from which they are produced are also depicted.
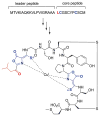
Fig. 33. Select examples of the linear description of RiPPs
The linear description is shown above each chemical structure. The 3–4 character descriptors are shown in bold red font, the amino acids to which they refer are shown in non-bold red font. Abu, 2-aminobutyric acid.
Fig. 34
Additional examples of linear shorthand notation for RiPPs.
Similar articles
- Ribosomally synthesized and post-translationally modified peptide natural products: new insights into the role of leader and core peptides during biosynthesis.
Yang X, van der Donk WA. Yang X, et al. Chemistry. 2013 Jun 10;19(24):7662-77. doi: 10.1002/chem.201300401. Epub 2013 May 10. Chemistry. 2013. PMID: 23666908 Free PMC article. - Protein Engineering in Ribosomally Synthesized and Post-translationally Modified Peptides (RiPPs).
Do T, Link AJ. Do T, et al. Biochemistry. 2023 Jan 17;62(2):201-209. doi: 10.1021/acs.biochem.1c00714. Epub 2022 Jan 10. Biochemistry. 2023. PMID: 35006671 Free PMC article. Review. - Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era.
Hetrick KJ, van der Donk WA. Hetrick KJ, et al. Curr Opin Chem Biol. 2017 Jun;38:36-44. doi: 10.1016/j.cbpa.2017.02.005. Epub 2017 Mar 2. Curr Opin Chem Biol. 2017. PMID: 28260651 Free PMC article. Review. - New Insights into the Biosynthetic Logic of Ribosomally Synthesized and Post-translationally Modified Peptide Natural Products.
Ortega MA, van der Donk WA. Ortega MA, et al. Cell Chem Biol. 2016 Jan 21;23(1):31-44. doi: 10.1016/j.chembiol.2015.11.012. Cell Chem Biol. 2016. PMID: 26933734 Free PMC article. Review. - Expanding the Structural Space of Ribosomal Peptides: Autocatalytic N-Methylation in Omphalotin Biosynthesis.
Aldemir H, Gulder TAM. Aldemir H, et al. Angew Chem Int Ed Engl. 2017 Oct 23;56(44):13570-13572. doi: 10.1002/anie.201708456. Epub 2017 Sep 26. Angew Chem Int Ed Engl. 2017. PMID: 28949431
Cited by
- Facile Removal of Leader Peptides from Lanthipeptides by Incorporation of a Hydroxy Acid.
Bindman NA, Bobeica SC, Liu WR, van der Donk WA. Bindman NA, et al. J Am Chem Soc. 2015 Jun 10;137(22):6975-8. doi: 10.1021/jacs.5b04681. Epub 2015 Jun 1. J Am Chem Soc. 2015. PMID: 26006047 Free PMC article. - Chemical synthesis of the lantibiotic lacticin 481 reveals the importance of lanthionine stereochemistry.
Knerr PJ, van der Donk WA. Knerr PJ, et al. J Am Chem Soc. 2013 May 15;135(19):7094-7. doi: 10.1021/ja4014024. Epub 2013 May 3. J Am Chem Soc. 2013. PMID: 23621626 Free PMC article. - Easy and rapid binding assay for functional analysis of disulfide-containing peptides by a pull-down method using a puromycin-linker and a cell-free translation system.
Tanemura Y, Mochizuki Y, Kumachi S, Nemoto N. Tanemura Y, et al. Biology (Basel). 2015 Mar 2;4(1):161-72. doi: 10.3390/biology4010161. Biology (Basel). 2015. PMID: 25738808 Free PMC article. - Biocatalytic synthesis of peptidic natural products and related analogues.
Liu D, Rubin GM, Dhakal D, Chen M, Ding Y. Liu D, et al. iScience. 2021 May 4;24(5):102512. doi: 10.1016/j.isci.2021.102512. eCollection 2021 May 21. iScience. 2021. PMID: 34041453 Free PMC article. Review. - novPTMenzy: a database for enzymes involved in novel post-translational modifications.
Khater S, Mohanty D. Khater S, et al. Database (Oxford). 2015 Apr 29;2015:bav039. doi: 10.1093/database/bav039. Print 2015. Database (Oxford). 2015. PMID: 25931459 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM039296/GM/NIGMS NIH HHS/United States
- BBS/E/J/00000607/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- DP2 OD008463/OD/NIH HHS/United States
- R01 AI078921/AI/NIAID NIH HHS/United States
- R01 GM088274/GM/NIGMS NIH HHS/United States
- R01 GM097142/GM/NIGMS NIH HHS/United States
- R01 AI022931/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources