Epigenetic inheritance mediated by histone lysine methylation: maintaining transcriptional states without the precise restoration of marks? - PubMed (original) (raw)
Review
Epigenetic inheritance mediated by histone lysine methylation: maintaining transcriptional states without the precise restoration of marks?
Chang Huang et al. Philos Trans R Soc Lond B Biol Sci. 2013.
Abstract
'Epigenetics' has been defined as the study of 'mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence'. Chromatin modifications are major carriers of epigenetic information that both reflect and affect the transcriptional states of underlying genes. Several histone modifications are key players that are responsible for classical epigenetic phenomena. However, the mechanisms by which cells pass their histone modifications to daughter cells through mitotic division remain to be unveiled. Here, we review recent progress in the field and conclude that epigenetic modifications are not precisely maintained at a near-mononucleosome level of precision. We also suggest that transcription repression may be maintained by a buffer system that can tolerate a certain degree of fluctuation in repressive histone modification levels. This buffer system protects the repressed genes from potential improper derepression triggered by chromatin modification-level fluctuation resulting from cellular events, such as the cell-cycle-dependent dilution of the chromatin modifications and local responses to environmental cues.
Figures
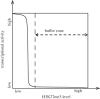
Figure 1.
A buffer model for mitotic inheritance of histone modifications. Bimodal relationship between transcriptional activity and the levels of repressive histone marks creates a buffer zone that allows the fluctuation of repressive histone modifications without sacrificing the faithful maintenance of target gene repression. H3K27me3 was used as a representative example for repressive histone marks.
Similar articles
- The C-terminus of histone H2B is involved in chromatin compaction specifically at telomeres, independently of its monoubiquitylation at lysine 123.
Wang CY, Hua CY, Hsu HE, Hsu CL, Tseng HY, Wright DE, Hsu PH, Jen CH, Lin CY, Wu MY, Tsai MD, Kao CF. Wang CY, et al. PLoS One. 2011;6(7):e22209. doi: 10.1371/journal.pone.0022209. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829450 Free PMC article. - Chromatin assembly factor 1 interacts with histone H3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair.
Zhou H, Madden BJ, Muddiman DC, Zhang Z. Zhou H, et al. Biochemistry. 2006 Mar 7;45(9):2852-61. doi: 10.1021/bi0521083. Biochemistry. 2006. PMID: 16503640 - Sir3 and epigenetic inheritance of silent chromatin in Saccharomyces cerevisiae.
Motwani T, Poddar M, Holmes SG. Motwani T, et al. Mol Cell Biol. 2012 Jul;32(14):2784-93. doi: 10.1128/MCB.06399-11. Epub 2012 May 14. Mol Cell Biol. 2012. PMID: 22586263 Free PMC article. - Histone variants and epigenetic inheritance.
Yuan G, Zhu B. Yuan G, et al. Biochim Biophys Acta. 2013 Mar-Apr;1819(3-4):222-229. doi: 10.1016/j.bbagrm.2011.06.007. Biochim Biophys Acta. 2013. PMID: 24459724 Review. - The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae.
Rusche LN, Kirchmaier AL, Rine J. Rusche LN, et al. Annu Rev Biochem. 2003;72:481-516. doi: 10.1146/annurev.biochem.72.121801.161547. Epub 2003 Mar 27. Annu Rev Biochem. 2003. PMID: 12676793 Review.
Cited by
- Epigenetic Research in Stem Cell Bioengineering-Anti-Cancer Therapy, Regenerative and Reconstructive Medicine in Human Clinical Trials.
Dompe C, Janowicz K, Hutchings G, Moncrieff L, Jankowski M, Nawrocki MJ, Józkowiak M, Mozdziak P, Petitte J, Shibli JA, Dyszkiewicz-Konwińska M, Bruska M, Piotrowska-Kempisty H, Kempisty B, Nowicki M. Dompe C, et al. Cancers (Basel). 2020 Apr 21;12(4):1016. doi: 10.3390/cancers12041016. Cancers (Basel). 2020. PMID: 32326172 Free PMC article. Review. - Mechanisms of epigenetic memory and addiction.
Tuesta LM, Zhang Y. Tuesta LM, et al. EMBO J. 2014 May 16;33(10):1091-103. doi: 10.1002/embj.201488106. Epub 2014 Apr 28. EMBO J. 2014. PMID: 24778453 Free PMC article. Review. - Epigenetic inheritance systems contribute to the evolution of a germline.
Lachmann M, Libby E. Lachmann M, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Aug 19;371(1701):20150445. doi: 10.1098/rstb.2015.0445. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27431523 Free PMC article. - Recognition of H3K9 methylation by GLP is required for efficient establishment of H3K9 methylation, rapid target gene repression, and mouse viability.
Liu N, Zhang Z, Wu H, Jiang Y, Meng L, Xiong J, Zhao Z, Zhou X, Li J, Li H, Zheng Y, Chen S, Cai T, Gao S, Zhu B. Liu N, et al. Genes Dev. 2015 Feb 15;29(4):379-93. doi: 10.1101/gad.254425.114. Epub 2015 Jan 30. Genes Dev. 2015. PMID: 25637356 Free PMC article.
References
- Russo VEA, Martienssen RA, Riggs AD. 1996. Introduction. In Epigenetic mechanisms of gene regulation, Russo VEA, Martienssen RA, Riggs AD. (eds), pp. 1–4 New York, NY: Cold Spring Harbor Laboratory Press
- Allis CD, Jenuwein T, Reinberg D. 2006. Overview and concepts. In Epigenetics, Allis CD, Jenuwein T, Reinberg D. (eds), pp. 23–56 New York: Cold Spring Harbor Laboratory
- Holliday R, Pugh JE. 1975. DNA modification mechanisms and gene activity during development. Science 187, 226–23210.1126/science.187.4173.226 (doi:10.1126/science.187.4173.226) - DOI - DOI - PubMed
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277, 1996–200010.1126/science.277.5334.1996 (doi:10.1126/science.277.5334.1996) - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases