A measure of the broad substrate specificity of enzymes based on 'duplicate' catalytic residues - PubMed (original) (raw)
Comparative Study
A measure of the broad substrate specificity of enzymes based on 'duplicate' catalytic residues
Sandeep Chakraborty et al. PLoS One. 2012.
Abstract
The ability of an enzyme to select and act upon a specific class of compounds with unerring precision and efficiency is an essential feature of life. Simultaneously, these enzymes often catalyze the reaction of a range of similar substrates of the same class, and also have promiscuous activities on unrelated substrates. Previously, we have established a methodology to quantify promiscuous activities in a wide range of proteins. In the current work, we quantitatively characterize the active site for the ability to catalyze distinct, yet related, substrates (BRASS). A protein with known structure and active site residues provides the framework for computing 'duplicate' residues, each of which results in slightly modified replicas of the active site scaffold. Such spatial congruence is supplemented by Finite difference Poisson Boltzmann analysis which filters out electrostatically unfavorable configurations. The congruent configurations are used to compute an index (BrassIndex), which reflects the broad substrate profile of the active site. We identify an acetylhydrolase and a methyltransferase as having the lowest and highest BrassIndex, respectively, from a set of non-homologous proteins extracted from the Catalytic Site Atlas. The acetylhydrolase, a regulatory enzyme, is known to be highly specific for platelet-activating factor. In the methyltransferase (PDB: 1QAM), various combinations of glycine (Gly38/40/42), asparagine (Asn101/11) and glutamic acid (Glu59/36) residues having similar spatial and electrostatic profiles with the specified scaffold (Gly38, Asn101 and Glu59) exemplifies the broad substrate profile such an active site may provide. 'Duplicate' residues identified by relaxing the spatial and/or electrostatic constraints can be the target of directed evolution methodologies, like saturation mutagenesis, for modulating the substrate specificity of proteins.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
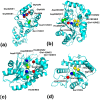
Figure 1. Active sites of proteins with the highest and lowest BrassIndex.
(a) rRNA Methyltransferase (PDBid:1QAM): This protein has the highest BrassIndex, as can be seen by the presence of various similar residues in close proximity, that results in electrostatically similar scaffolds as well (Table 2). (b) Palmitoyl protein thioesterase 1 (PPT1) (PDBid:1EH5): Protein with the next highest BrassIndex. We hypothesize that a ‘replica’ catalytic triad consisting of Asp288 exists and is congruent to the known catalytic triad (His289, Asp233, Ser115) (Table 3). (c) Palmitoyl protein thioesterase 2 (PPT2) (PDBid:1PJA): PPT2 has a 26% similarity with PPT1, but has a non-redundant role in the cell. The absence of supporting residues could be a possible reason why PPT2 is unable to act upon all compounds (particularly those with have bulky head groups) which PPT1 catalyzes. (d) Platelet-activating acetylhydrolase (PDBid:1BWP): This protein has the lowest BrassIndex, which is due to the absence of ‘duplicate’ residues in the proximity of the core active site residues (Table 5). This implies that this protein has high specificity, a fact that has been noted in .
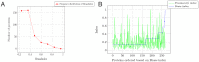
Figure 2. Statistics of BrassIndex on the population: (a) Frequency distribution of BrassIndex.
It can be seen that most proteins are highly specific (low BrassIndex), and the number of proteins with high specificity drops exponentially. (b) Lack of correlation between promiscuity and substrate specificity (Brass) indices: As expected, there is no correlation between promiscuity (defined as the ability to catalyze reactions distinct from the one the protein has evolved to perform, but using the same active site scaffold) and the ability of enzymes to catalyze the reaction of different, but related, compounds using the same catalytic mechanism (broad substrate specificity). The promiscuity indices are computed as described in .
Similar articles
- Shining a light on enzyme promiscuity.
Copley SD. Copley SD. Curr Opin Struct Biol. 2017 Dec;47:167-175. doi: 10.1016/j.sbi.2017.11.001. Epub 2017 Nov 21. Curr Opin Struct Biol. 2017. PMID: 29169066 Review. - An automated flow for directed evolution based on detection of promiscuous scaffolds using spatial and electrostatic properties of catalytic residues.
Chakraborty S. Chakraborty S. PLoS One. 2012;7(7):e40408. doi: 10.1371/journal.pone.0040408. Epub 2012 Jul 11. PLoS One. 2012. PMID: 22811760 Free PMC article. - The evolution of multiple active site configurations in a designed enzyme.
Hong NS, Petrović D, Lee R, Gryn'ova G, Purg M, Saunders J, Bauer P, Carr PD, Lin CY, Mabbitt PD, Zhang W, Altamore T, Easton C, Coote ML, Kamerlin SCL, Jackson CJ. Hong NS, et al. Nat Commun. 2018 Sep 25;9(1):3900. doi: 10.1038/s41467-018-06305-y. Nat Commun. 2018. PMID: 30254369 Free PMC article. - Bioinformatics and molecular modeling in chemical enzymology. Active sites of hydrolases.
Varfolomeev SD, Uporov IV, Fedorov EV. Varfolomeev SD, et al. Biochemistry (Mosc). 2002 Oct;67(10):1099-108. doi: 10.1023/a:1020907122341. Biochemistry (Mosc). 2002. PMID: 12460108 Review. - Unusual commonality in active site structural features of substrate promiscuous and specialist enzymes.
Thakur D, Pandit SB. Thakur D, et al. J Struct Biol. 2022 Mar;214(1):107835. doi: 10.1016/j.jsb.2022.107835. Epub 2022 Jan 31. J Struct Biol. 2022. PMID: 35104611
Cited by
- Multiple transport-active binding sites are available for a single substrate on human P-glycoprotein (ABCB1).
Chufan EE, Kapoor K, Sim HM, Singh S, Talele TT, Durell SR, Ambudkar SV. Chufan EE, et al. PLoS One. 2013 Dec 5;8(12):e82463. doi: 10.1371/journal.pone.0082463. eCollection 2013. PLoS One. 2013. PMID: 24349290 Free PMC article. - Oligomeric state regulated trafficking of human platelet-activating factor acetylhydrolase type-II.
Monillas ES, Caplan JL, Thévenin AF, Bahnson BJ. Monillas ES, et al. Biochim Biophys Acta. 2015 May;1854(5):469-75. doi: 10.1016/j.bbapap.2015.02.007. Epub 2015 Feb 20. Biochim Biophys Acta. 2015. PMID: 25707358 Free PMC article. - Intra-site differential inhibition of multi-specific enzymes.
Cappiello M, Balestri F, Moschini R, Mura U, Del-Corso A. Cappiello M, et al. J Enzyme Inhib Med Chem. 2020 Dec;35(1):840-846. doi: 10.1080/14756366.2020.1743988. J Enzyme Inhib Med Chem. 2020. PMID: 32208768 Free PMC article. Review. - Promiscuous Ribozymes and Their Proposed Role in Prebiotic Evolution.
Janzen E, Blanco C, Peng H, Kenchel J, Chen IA. Janzen E, et al. Chem Rev. 2020 Jun 10;120(11):4879-4897. doi: 10.1021/acs.chemrev.9b00620. Epub 2020 Feb 3. Chem Rev. 2020. PMID: 32011135 Free PMC article. Review. - Biological messiness vs. biological genius: Mechanistic aspects and roles of protein promiscuity.
Atkins WM. Atkins WM. J Steroid Biochem Mol Biol. 2015 Jul;151:3-11. doi: 10.1016/j.jsbmb.2014.09.010. Epub 2014 Sep 12. J Steroid Biochem Mol Biol. 2015. PMID: 25218442 Free PMC article. Review.
References
- Nelson DL, Cox MM (2008) Lehninger’s Principles of Biochemistry. W. H. Freeman, fifth edition.
- Bone R, Silen JL, Agard DA (1989) Structural plasticity broadens the specificity of an engineered protease. Nature 339: 191–195. - PubMed
- Huse M, Kuriyan J (2002) The conformational plasticity of protein kinases. Cell 109: 275–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This work was funded by the Tata Institute of Fundamental Research (Department of Atomic Energy), and the Department of Science and Technology (JC Bose Award Grant). BA extends gratitude to the Icelandic National Research Council (RANNIS) and the University of Iceland Research Found for supporting the project financially. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources