Transcription elongation factor Tcea3 regulates the pluripotent differentiation potential of mouse embryonic stem cells via the Lefty1-Nodal-Smad2 pathway - PubMed (original) (raw)
Young Cha, Chun-Hyung Kim, Hee-Jin Ahn, Dohoon Kim, Sanghyeok Ko, Kyeoung-Hwa Kim, Mi-Yoon Chang, Jong-Hyun Ko, Yoo-Sun Noh, Yong-Mahn Han, Jonghwan Kim, Jihwan Song, Jin Young Kim, Paul J Tesar, Robert Lanza, Kyung-Ah Lee, Kwang-Soo Kim
Affiliations
- PMID: 23169579
- PMCID: PMC3572291
- DOI: 10.1002/stem.1284
Transcription elongation factor Tcea3 regulates the pluripotent differentiation potential of mouse embryonic stem cells via the Lefty1-Nodal-Smad2 pathway
Kyung-Soon Park et al. Stem Cells. 2013 Feb.
Abstract
Self-renewal and pluripotency are hallmark properties of pluripotent stem cells, including embryonic stem cells (ESCs) and iPS cells. Previous studies revealed the ESC-specific core transcription circuitry and showed that these core factors (e.g., Oct3/4, Sox2, and Nanog) regulate not only self-renewal but also pluripotent differentiation. However, it remains elusive how these two cell states are regulated and balanced during in vitro replication and differentiation. Here, we report that the transcription elongation factor Tcea3 is highly enriched in mouse ESCs (mESCs) and plays important roles in regulating the differentiation. Strikingly, altering Tcea3 expression in mESCs did not affect self-renewal under nondifferentiating condition; however, upon exposure to differentiating cues, its overexpression impaired in vitro differentiation capacity, and its knockdown biased differentiation toward mesodermal and endodermal fates. Furthermore, we identified Lefty1 as a downstream target of Tcea3 and showed that the Tcea3-Lefty1-Nodal-Smad2 pathway is an innate program critically regulating cell fate choices between self-replication and differentiation commitment. Together, we propose that Tcea3 critically regulates pluripotent differentiation of mESCs as a molecular rheostat of Nodal-Smad2/3 signaling.
Copyright © 2012 AlphaMed Press.
Figures
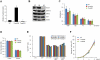
Figure 1. Transcription elongation factor Tcea3 is predominantly expressed in undifferentiated mESCs, oocytes, and early embryos
(A) Membrane array analysis was performed using total RNA samples from mouse oocytes, mESCs, MEFs, and NIH3T3 cells. The spots indicated by arrows correspond to Tcea3, Gbx2 and Tbx33. (B) RT-PCR analysis reveals that Tcea3 is expressed in mouse oocytes and ESCs but not in MEF and NIH3T3 cells. (C) Relative expression of Tcea1, Tcea2 and Tcea3 in mESCs and oocytes, as analyzed by real-time RT-PCR. (D) Analysis of mouse tissue-specific expression of Tcea1, Tcea2 and Tcea3 transcripts by RT-PCR of total RNA prepared from the indicated tissues. (E) Expression analyses of Tcea3 by RT-PCR from total RNA and immunoblotting of whole cell extracts from ESCs, EBs and in vitro differentiated cells at day 3 and 6, following LIF withdrawal and RA addition (RA-d3, RA-d6). GAPDH and α-tubulin were used as loading controls for RT-PCR or Western blot analysis, respectively. (F) Expression of Tcea3 in oocytes, fertilized eggs, and early stage mouse embryos by immunocytochemical staining. All values are means ± s.d. from at least triplicate experiments. ** Indicates highly significant (P<0.01) results based on Student's T-test analyses. Abbreviations: GV, germinal vesicle; PN, pronucleus; MO, morula; BL, blastocyst.
Figure 2. Altered expression levels of Tcea3 do not affect self-renewal of mESCs
(A) Tcea3 transcript levels were analyzed by realtime RT-PCR. (B) Protein expression of Tcea3, Oct4, and p-Stat3 was analyzed by immunoblot using cell extracts from WT, Tcea3 OE, and KD mESCs. (C) WT, Tcea3 OE and Tcea3 KD mESCs were maintained in different concentrations of LIF for 5 days and AP activity was measured. (D) 1st EBs from indicated cells were dissociated into single cells and re-seeded at a density of 1 × 106 cells/ml in the same medium. The number of 2nd EB colonies was counted under light microscope. (E) GFP-positive (GFP+) mESCs were mixed at a ratio of 1:1 with GFP-negative (GFP-) WT, Tcea3 OE, and Tcea3 KD cells, respectively. The GFP+/GFP- ratios were measured at each passage. (F) Cell proliferation of WT, Tcea3 OE and Tcea3 KD mESCs was analyzed by counting cell number every 2 days under ESC culture condition. All values are means ± s.d. from at least triplicate experiments. * indicates significant (P<0.05) results based on Student's T-test analyses.
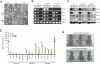
Figure 3. Altered expression of Tcea3 influences multi-lineage differentiation potential of mESCs both in vitro and in vivo
(A) In vitro differentiation was induced by removing LIF and adding RA to WT, Tcea3 OE, and KD mESCs. Cells were examined at day 0 (D0), day 2 (D2) or day 4 (D4) following in vitro differentiation. Scale bar = 100 μm. (B) RT-PCR analysis of Tcea3, Oct4, Sox2 and Nanog expression during in vitro differentiation of WT, Tcea3 OE, and KD mESCs. (C) Immunoblot analysis of Tcea3, Oct4, Sox2 and Nanog expression during in vitro differentiation of WT, Tcea3 OE, and KD mESCs. (D) WT, Tcea3 OE, and KD mESCs differentiated for 4 days (RA-d4) were analyzed for the expression of markers representing ectoderm, mesoderm and endoderm by real-time RT-PCR. The expression level of each gene was shown as relative value following normalization against that of the glyceraldehyde 3-phosphate dehydrogenase (Gapdh) gene. (E) WT, Tcea3 OE and Tcea3 KD cells were injected into NOD/SCID mice and teratoma development was examined. Teratoma formation of Tcea3 OE cells was compared with that of WT cells 8 weeks after injection and teratoma formation of Tcea3 KD cells was compared with that of WT cells 4 weeks after injection. This teratoma analysis was repeated twice with identical results (data not shown). All values are means ± s.d. from at least triplicate experiments. * indicates significant(P<0.05) and ** highly significant (p<0.01) results based on ANOVA analyses following the Scheffe test.
Figure 4. Tcea3 is a component of RNA polymerase II transcription complex and regulates expression of Lefty1 in mESCs
(A) Agarose gel analysis of Tcea3 binding proteins from mESCs. mESC total cell extracts were used as “prey” and GST fused Tcea3 were used as “bait” for the GST pull down assay. (B) List of representative proteins identified as protein binding partners of Tcea3 by the mass spectrometric analysis of peptides extracted from four agarose bands of (A). (C) Scatter plots of cDNA microarray analysis of Tcea3 OE mESCs revealed that Lefty1 expression is most robustly upregulated (R2 = correlation coefficients).
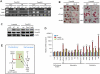
Figure 5. Lefty1 is a downstream target gene of Tcea3
(A) Real-time RT-PCR analysis confirms the microarray results. (B) Real-time RT-PCR analysis shows that transient transfection of Tcea3_-expressing vector (Tcea3 OE (T)) dramatically induced Tcea3 (left) and Lefty1 (right) transcript expression and that co-transfection of Tcea3_-specific siRNA reduced them. Non-specific siRNA (siNS) was transfected as control. The error bars correspond to three replicates (n=3) and show the mean ± s.d. (C) Lefty1 expression in Tcea3 KD mESCs compared with that of WT mESCs by qRT-PCR (left) and immunoblotting analysis (right). (D) Immunoblotting results of p-Smad2 in Tcea3 OE and WT mESCs at 0 (ES) or 4 days (RA-d4) during in vitro differentiation. β-actin was used as loading control. (E) Expression levels of Lefty1 and p-Smad2 were analyzed by immunoblotting after siRNA-mediated transient knockdown of Tcea3 in WT mESCs. All values are means ± s.d. from at least triplicate experiments. * indicates significant (P<0.05) and ** highly significant (p<0.01) results based on Student's T-test analyses. Abbreviation: siNS, non-specific siRNA; si_Tcea3, siRNA targeting to Tcea3; si_Lefty1, siRNA targeting to Lefty1.
Figure 6. Tcea3 controls the in vitro differentiation potential of mESCs by regulating the Lefty1 expression
(A) Tcea3 OE mESCs were transfected with si_Tcea3_ or si_Lefty1_ and transcript levels of Tcea3 and Lefty1 were analyzed by RT-PCR either at 0 or 2 days following in vitro differentiation (left). Tcea3 KD mESCs were transfected with Tcea3 or Lefty1 expressing plasmid and transcript levels were analyzed by RT-PCR (right). (B) Tcea3 OE mESCs were transfected with si_Tcea3_ or si_Lefty1_ and Tcea3 KD mESCs were transfected with Tcea3 or Lefty1 expressing plasmid. Differentiation was analyzed by morphological changes and AP staining. (C) Tcea3 OE mESCs were transfected with siRNA targeting Tcea3 or Lefty1 and Tcea3 KD mESCs were transfected with Tcea3 or Lefty1 expressing plasmids for 24 hr. Levels of pSmad2 were analyzed by immunoblotting. (D) Tcea3 KD mESCs were differentiated for 2 days by removing LIF and adding RA in the presence or absence of SB431542 (20 μM) and the expression of self-renewal factors or mesoendoderm marker genes were analyzed by RT-PCR. (E) A schematic diagram depicting the proposed role of the Tcea3-Lefty1_-Nodal-Smad2 pathway controlling cell fate choices between self-renewal and differentiation commitment. Proper expression levels of Tcea3 appear to be critical for balancing the transition between these two cell fates. In addition, our model suggests that Tcea3 and Lefty1 importantly link these two cell fates by being regulated by core transcription factors and inhibiting Nodal signaling (see text). All values are means ± s.d. from at least triplicate experiments. ** indicates highly significant (P<0.01) results based on ANOVA analyses following the Scheffe test. Abbreviation: siNS, non-specific siRNA; si_T, siRNA targeting Tcea3; si_L_, siRNA targeting to Lefty1.
Similar articles
- Lefty1 and lefty2 control the balance between self-renewal and pluripotent differentiation of mouse embryonic stem cells.
Kim DK, Cha Y, Ahn HJ, Kim G, Park KS. Kim DK, et al. Stem Cells Dev. 2014 Mar 1;23(5):457-66. doi: 10.1089/scd.2013.0220. Epub 2013 Nov 28. Stem Cells Dev. 2014. PMID: 24147624 Free PMC article. - Nodal signaling regulates the bone morphogenic protein pluripotency pathway in mouse embryonic stem cells.
Galvin KE, Travis ED, Yee D, Magnuson T, Vivian JL. Galvin KE, et al. J Biol Chem. 2010 Jun 25;285(26):19747-56. doi: 10.1074/jbc.M109.077347. Epub 2010 Apr 28. J Biol Chem. 2010. PMID: 20427282 Free PMC article. - Tcea3 regulates the vascular differentiation potential of mouse embryonic stem cells.
Cha Y, Heo SH, Ahn HJ, Yang SK, Song JH, Suh W, Park KS. Cha Y, et al. Gene Expr. 2013;16(1):25-30. doi: 10.3727/105221613x13776146743343. Gene Expr. 2013. PMID: 24397209 Free PMC article. - Transforming growth factor-beta superfamily in mouse embryonic stem cell self-renewal.
Galvin-Burgess KE, Vivian JL. Galvin-Burgess KE, et al. Vitam Horm. 2011;87:341-65. doi: 10.1016/B978-0-12-386015-6.00035-4. Vitam Horm. 2011. PMID: 22127250 Review. - Roles of TGF-β family signals in the fate determination of pluripotent stem cells.
Itoh F, Watabe T, Miyazono K. Itoh F, et al. Semin Cell Dev Biol. 2014 Aug;32:98-106. doi: 10.1016/j.semcdb.2014.05.017. Epub 2014 Jun 6. Semin Cell Dev Biol. 2014. PMID: 24910449 Review.
Cited by
- Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis.
Urh K, Žlajpah M, Zidar N, Boštjančič E. Urh K, et al. Biomedicines. 2021 Feb 11;9(2):179. doi: 10.3390/biomedicines9020179. Biomedicines. 2021. PMID: 33670246 Free PMC article. - Novel function of E26 transformation-specific domain-containing protein ELK3 in lymphatic endothelial cells.
Park JI, Kim KS, Kong SY, Park KS. Park JI, et al. Oncol Lett. 2018 Jan;15(1):55-60. doi: 10.3892/ol.2017.7308. Epub 2017 Oct 31. Oncol Lett. 2018. PMID: 29375705 Free PMC article. - Regulation of hypoxia responses by flavin adenine dinucleotide-dependent modulation of HIF-1α protein stability.
Yang SJ, Park YS, Cho JH, Moon B, An HJ, Lee JY, Xie Z, Wang Y, Pocalyko D, Lee DC, Sohn HA, Kang M, Kim JY, Kim E, Park KC, Kim JA, Yeom YI. Yang SJ, et al. EMBO J. 2017 Apr 13;36(8):1011-1028. doi: 10.15252/embj.201694408. Epub 2017 Mar 9. EMBO J. 2017. PMID: 28279976 Free PMC article. - The transcription elongation factor TCEA3 promotes the activity of the myogenic regulatory factors.
Kazim N, Adhikari A, Davie J. Kazim N, et al. PLoS One. 2019 Jun 3;14(6):e0217680. doi: 10.1371/journal.pone.0217680. eCollection 2019. PLoS One. 2019. PMID: 31158246 Free PMC article. - An integrated systems biology approach identifies positive cofactor 4 as a factor that increases reprogramming efficiency.
Jo J, Hwang S, Kim HJ, Hong S, Lee JE, Lee SG, Baek A, Han H, Lee JI, Lee I, Lee DR. Jo J, et al. Nucleic Acids Res. 2016 Feb 18;44(3):1203-15. doi: 10.1093/nar/gkv1468. Epub 2016 Jan 5. Nucleic Acids Res. 2016. PMID: 26740582 Free PMC article.
References
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. - PubMed
- Sato N, Meijer L, Skaltsounis L, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. - PubMed
- Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–884. - PubMed
- Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC4 HL106627/HL/NHLBI NIH HHS/United States
- R01 NS084869/NS/NINDS NIH HHS/United States
- HL106627/HL/NHLBI NIH HHS/United States
- R21 MH087903/MH/NIMH NIH HHS/United States
- NS070577/NS/NINDS NIH HHS/United States
- R01 NS070577/NS/NINDS NIH HHS/United States
- MH087903/MH/NIMH NIH HHS/United States
- R33 MH087903/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials