Dnmt1-dependent DNA methylation is essential for photoreceptor terminal differentiation and retinal neuron survival - PubMed (original) (raw)
Dnmt1-dependent DNA methylation is essential for photoreceptor terminal differentiation and retinal neuron survival
K-D Rhee et al. Cell Death Dis. 2012.
Abstract
Epigenetic regulation of the genome is critical for the emergence of diverse cell lineages during development. To understand the role of DNA methylation during retinal network formation, we generated a mouse retinal-specific Dnmt1 deletion mutation from the onset of neurogenesis. In the hypomethylated Dnmt1-mutant retina, neural progenitor cells continue to proliferate, however, the cell cycle progression is altered, as revealed by an increased proportion of G1 phase cells. Despite production of all major retinal neuronal cell types in the Dnmt1-mutant retina, various postmitotic neurons show defective differentiation, including ectopic cell soma and aberrant dendritic morphologies. Specifically, the commitment of Dmnt1-deficient progenitors towards the photoreceptor fate is not affected by DNA hypomethylation, yet the initiation of photoreceptor differentiation is severely hindered, resulting in reduction and mislocalization of rhodopsin-expressing cells. In addition to compromised neuronal differentiation, Dnmt1 deficiency also leads to rapid cell death of photoreceptors and other types of neurons in the postnatal retina. These results indicate that Dnmt1-dependent DNA methylation is critical for expansion of the retinal progenitor pool, as well as for maturation and survival of postmitotic neurons.
Figures
Figure 1
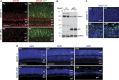
Effects of retinal-specific deletion of Dnmt1 by Chx10-Cre. (a) Confocal micrographs of retinas co-labeled for Dnmt1 (red) and GFP (green) at P3. The Dnmt1 cKO retina shows reduced Dnmt1 protein expression compared with the heterozygous control. White arrows point to cells co-expressing Dnmt1 and Cre-GFP. (b) Western blots show reduction of Dnmt1 protein in two individual P3 Dnmt1 cKO mutant retinas that also express high levels of IAP. (c) Confocal images of the outer retinas co-labeled for DAPI and IAP at P3 and P7. Dnmt1 cKO mutant retinas contain IAP+ cells with altered nuclear morphology. (d) DAPI labeling shows progressive thinning of __Dnmt1-_mutant retina at P3, P7 and P30. Scale bars, (a) 50 _μ_m, (c) 10 _μ_m, (d) P3, 100 _μ_m; P7 and P30, 50 _μ_m. ac, amacrine cells; gcl, ganglion cell layer; inl, inner nuclear layer; ipl, inner plexiform layer; vs, ventricular surface; vz, ventricular zone
Figure 2
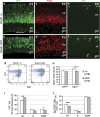
Dnmt1 deletion alters progenitor distribution and cell cycle progression. (a–f) Immunolabeling of retinas show distribution of progenitor markers GFP (a and b) and proliferating cell nuclear antigen (PCNA, c and d) at P3, and M-phase marker PH3 (e and f) at P0. Arrow in f indicates an ectopic PH3+ cell. Scale bar, (a) for (a–f) 50 _μ_m. gcl, ganglion cell layer; ipl, inner plexiform layer; vz, ventricular zone. (g–j) Flow cytometric analyses of cell markers and cell cycle properties at P2. (g) Representative flow cytometry profiles for GFP and cyclin-dependent protein kinase inhibitor p27Kip1. (h) Fluorescence-activated cell sorting quantification of GFP+ cells and p27Kip1+ cells among total cells. (i) Cell cycle distributions of GFP+ progenitors. (j) Quantification of p27Kip1+ GFP+ cells among GFP+ cells in different cell cycle phases. Numbers of individual retinas analyzed in (h–j) are indicated in h. *P<0.05, **P<0.01, ***P<0.001
Figure 3
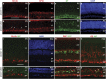
Effects of Dnmt1 deletion on retinal cell type production. (a–j) Immunolabeling of early born cell types in the neonatal retina. Cell marker labeling for RGCs (a and b), amacrine cells (c and d), horizontal cells (e and f) are presented. i and j shows co-labeling of Brn3a and IAP. White arrows (b and j) point to mislocalized RGCs. DAPI staining (g and h) shows retinal laminar structures at P3. (k–p) Confocal images of P30 retinas for late-born retinal cell types. Cell marker labeling for bipolar cells (m and n) and Muller cells (o and p) are presented. Arrows in n point to rod bipolar cells that are double labeled for Cre-GFP and protein kinase C-α (PKC_α_), and p indicate mutant Muller cell double labeled for IAP and glutamine synthetase (GS). Scale bars, a for a–h, 100 _μ_m; i for i–p, 50 _μ_m. gcl, ganglion cell layer; inl, inner nuclear layer; vz, ventricular zone
Figure 4
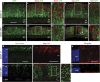
Effects of Dnmt1 mutation on photoreceptor commitment and differentiation. (a–f') Confocal images of co-immunolabeling at P3 for Otx2 and GFP (a–d'), or Crx and GFP (e–f'). c', d' and e', f' show twofold magnification of framed areas in c, d and e, f, respectively. The Crx+ cells in control (e and e') and in Cre− regions of the mutant (f and f') are located near the ventricular surface; but __Dnmt1-_deficient Crx+ cells are distributed throughout vz (f and f'). Arrows indicate co-labeled GFP+ Crx+ cells. (g and h) Immunolabeling for recoverin and DAPI at P3. (i, j and j') Confocal images of co-labeling for Rho (red) and IAP (green) at P3. A lack of Rho+ cells is seen in _Dnmt1_-deficient regions expressing IAP (j). A rare Rho and IAP co-labeled cell framed in j is shown in j' at twofold magnification. (k and l) Confocal images of immunolabeling for M-opsin at P3. Scale bars, a for a–d, e and f, g for g and h, i for i and j, 50 _μ_m; k for k and l, 25 _μ_m. gcl, ganglion cell layer; ipl, inner plexiform layer; ventricular surface; vz, ventricular zone
Figure 5
Dnmt1 ablation causes DNA hypomethylation and cell death in the postnatal retina. (a) DNA methylation patterns at Rhodopsin and M-opsin promoters as determined by bisulfide sequencing. Significantly reduced CpG dinucleotide methylation is detected at P3 in the mutant retina. Columns correspond to CpG sites with the relative location from the transcription-starting sites (arrows), while rows correspond to sequenced clones. Solid circles indicate methylated CpG; open circles indicate unmethylated CpG. (b and c) Immunolabeling for activated caspase 3 shows increased apoptosis in Dnmt1 cKO retinas at P3. Note that Casp3+ cells are distributed throughout the mutant retina. (d and e) Co-labeling of DAPI and Rho at P5. f and g represent magnified regions framed in d and e to show condensed nuclei (arrows) present in mutant cells. Scale bars, b for b and c, d for d and e, 50 _μ_m; f for f and g, 10 _μ_m. gcl, ganglion cell layer; inl, inner nuclear layer; ipl, inner plexiform layer; vz, ventricular zone
Similar articles
- Dnmt1, Dnmt3a and Dnmt3b cooperate in photoreceptor and outer plexiform layer development in the mammalian retina.
Singh RK, Mallela RK, Hayes A, Dunham NR, Hedden ME, Enke RA, Fariss RN, Sternberg H, West MD, Nasonkin IO. Singh RK, et al. Exp Eye Res. 2017 Jun;159:132-146. doi: 10.1016/j.exer.2016.11.014. Epub 2016 Nov 16. Exp Eye Res. 2017. PMID: 27865785 - DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals.
Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, Trumpp A, Poon C, Wilson CB, Jaenisch R. Fan G, et al. J Neurosci. 2001 Feb 1;21(3):788-97. doi: 10.1523/JNEUROSCI.21-03-00788.2001. J Neurosci. 2001. PMID: 11157065 Free PMC article. - Distinct nuclear localization patterns of DNA methyltransferases in developing and mature mammalian retina.
Nasonkin IO, Lazo K, Hambright D, Brooks M, Fariss R, Swaroop A. Nasonkin IO, et al. J Comp Neurol. 2011 Jul 1;519(10):1914-30. doi: 10.1002/cne.22613. J Comp Neurol. 2011. PMID: 21452232 - Epigenetic regulation in the commitment of progenitor cells during retinal development and regeneration.
Yin W, Mao X, Xu M, Chen M, Xue M, Su N, Yuan S, Liu Q. Yin W, et al. Differentiation. 2023 Jul-Aug;132:51-58. doi: 10.1016/j.diff.2023.04.002. Epub 2023 Apr 12. Differentiation. 2023. PMID: 37069005 Review. - Epigenetic regulation of retinal development.
Raeisossadati R, Ferrari MFR, Kihara AH, AlDiri I, Gross JM. Raeisossadati R, et al. Epigenetics Chromatin. 2021 Feb 9;14(1):11. doi: 10.1186/s13072-021-00384-w. Epigenetics Chromatin. 2021. PMID: 33563331 Free PMC article. Review.
Cited by
- DNA Methyltransferase 1 (DNMT1) Function Is Implicated in the Age-Related Loss of Cortical Interneurons.
Hahn A, Pensold D, Bayer C, Tittelmeier J, González-Bermúdez L, Marx-Blümel L, Linde J, Groß J, Salinas-Riester G, Lingner T, von Maltzahn J, Spehr M, Pieler T, Urbach A, Zimmer-Bensch G. Hahn A, et al. Front Cell Dev Biol. 2020 Jul 22;8:639. doi: 10.3389/fcell.2020.00639. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32793592 Free PMC article. - Nuclear NAD+-biosynthetic enzyme NMNAT1 facilitates development and early survival of retinal neurons.
Sokolov D, Sechrest ER, Wang Y, Nevin C, Du J, Kolandaivelu S. Sokolov D, et al. Elife. 2021 Dec 8;10:e71185. doi: 10.7554/eLife.71185. Elife. 2021. PMID: 34878972 Free PMC article. - Epigenetics in ocular diseases.
Liu MM, Chan CC, Tuo J. Liu MM, et al. Curr Genomics. 2013 May;14(3):166-72. doi: 10.2174/1389202911314030002. Curr Genomics. 2013. PMID: 24179439 Free PMC article. - DNA Methylation-Dependent Dysregulation of GABAergic Interneuron Functionality in Neuropsychiatric Diseases.
Linde J, Zimmer-Bensch G. Linde J, et al. Front Neurosci. 2020 Sep 16;14:586133. doi: 10.3389/fnins.2020.586133. eCollection 2020. Front Neurosci. 2020. PMID: 33041771 Free PMC article. Review. - Regulation of Opsin Gene Expression by DNA Methylation and Histone Acetylation.
Song J, VanBuskirk JA, Merbs SL. Song J, et al. Int J Mol Sci. 2022 Jan 26;23(3):1408. doi: 10.3390/ijms23031408. Int J Mol Sci. 2022. PMID: 35163334 Free PMC article.
References
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. - PubMed
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 (Suppl:245–254. - PubMed
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
- Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1:11–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 EY019052/EY/NEI NIH HHS/United States
- EY000331/EY/NEI NIH HHS/United States
- EY019052/EY/NEI NIH HHS/United States
- R01 EY012270/EY/NEI NIH HHS/United States
- P30 EY000331/EY/NEI NIH HHS/United States
- EY12270/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases