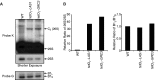Las1 interacts with Grc3 polynucleotide kinase and is required for ribosome synthesis in Saccharomyces cerevisiae - PubMed (original) (raw)
Las1 interacts with Grc3 polynucleotide kinase and is required for ribosome synthesis in Saccharomyces cerevisiae
Christopher D Castle et al. Nucleic Acids Res. 2013 Jan.
Abstract
Ribosome biogenesis is a multi-step process that couples cell growth with cell proliferation. Although several large-scale analysis of pre-ribosomal particles have identified numerous trans-acting factors involved in this process, many proteins involved in pre-rRNA processing and ribosomal subunit maturation have yet to be identified. Las1 was originally identified in Saccharomyces cerevisiae as a protein involved in cell morphogenesis. We previously demonstrated that the human homolog, Las1L, is required for efficient ITS2 rRNA processing and synthesis of the 60S ribosomal subunit. Here, we report that the functions of Las1 in ribosome biogenesis are also conserved in S. cerevisiae. Depletion of Las1 led to the accumulation of both the 27S and 7S rRNA intermediates and impaired the synthesis of the 60S subunit. We show that Las1 co-precipitates mainly with the 27S rRNA and associates with an Nsa1 and Rix1-containing pre-60S particle. We further identify Grc3 as a major Las1-interacting protein. We demonstrate that the kinase activity of Grc3 is required for efficient pre-rRNA processing and that depletion of Grc3 leads to rRNA processing defects similar to the ones observed in Las1-depleted cells. We propose that Las1 and Grc3 function together in a conserved mechanism to modulate rRNA processing and eukaryotic ribosome biogenesis.
Figures
Figure 1.
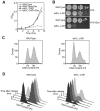
Las1 is required for cell cycle progression. (A) Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 24 h. Cells were then diluted into YPD with 10 µg/ml doxycycline and OD600 was measured at the indicated time points. (B) Wild type and tetO7-LAS1 strains were grown in YPD or YPD with 10 µg/ml doxycycline and then diluted to an OD600 of 0.05. Ten-fold serial dilutions were performed and 5 µl was plated on YPD plates with or without 10 µg/ml doxycycline and incubated at 30°C for 48 h. (C) Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline to log phase, stained with propidium iodide, and subjected to FACS analysis. (D) Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 24 h and cells were then arrested by the addition of α-factor. Cells were released into the cell cycle by washing with water and diluted into YPD with 10 µg/ml doxycycline. Indicated time points were collected and cells were stained with propidium iodide and analysed by FACS analysis.
Figure 2.
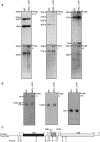
Las1 is required for pre-rRNA processing. (A) Northern blot analysis of steady-state pre-rRNAs. Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 12 h. Equal amounts of RNA were hybridized with probes specific for the indicated region of pre-rRNA. The position of each pre-rRNA is indicated on the left of each panel. Note that the signal detected with probe A above the 23S band corresponds to unspecific binding of the probe to the 25S rRNA. (B) Northern blot analysis of low molecular weight RNAs. Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 12 h. Equal amounts of total RNA were separated on an 8% denaturing acrylamide gel and hybridized with probes specific for the indicated region of pre-rRNA. The positions of each pre-rRNA are indicated on the left of each panel. (C) Schematic representation of the rRNA primary transcript in budding yeast. The position of the probes utilized for the northern blotting analyses in A and B are indicated with letters.
Figure 3.
Depletion of Las1 and Grc3 leads to the accumulation of a 26S rRNA intermediate. (A) Primer extension analysis. Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 12 h. Equal amounts of RNA were subjected to primer extension analysis using probes as indicated on the left side of each panel. The position of each pre-rRNA is indicated on the right of each panel. (B) Densitometry analysis of the relative 26S/25S and B1L/B1S ratios for the primer extension analysis in (A).
Figure 4.
Depletion of Las1 impairs 25S rRNA synthesis. (A) Pulse-chase analysis of newly synthesized pre-rRNAs. Wild type and tetO7-LAS1 strains were grown in YPD with 10 µg/ml doxycycline for 6 h. Cells were starved of methionine for 30 min and then pulse labeled with 3H-methyl-methionine for 1 min. Cells were then chased for the indicated time points with an excess of cold methionine and total RNA was harvested. 20 000 cpm was separated on a 1.2% agarose-formaldehyde denaturing gel, transferred to a nylon membrane, and visualized by autoradiography. (B) Pulse-chase analysis of newly synthesized small pre-rRNAs. Wild type and tetO7-LAS1 strains were grown in uracil-depleted media with 10 µg/ml doxycycline for 6 h. Cells were pulse labeled with [5,6-3H]-uracil for 1 min and then chased for the indicated time points with an excess of cold uracil and total RNA was harvested. Equal counts were loaded and separated on an 8% acrylamide-formaldehyde denaturing gel, transferred to a nylon membrane, and visualized by autoradiography.
Figure 5.
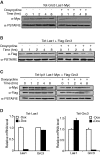
Las1 interacts with Grc3 polynucleotide kinase. (A) Mass spectrometry analysis of Las1-associated proteins. BY4741 and Las1-Myc strains were grown to OD600 of 0.6 and proteins were immunoprecipitated from lysates with an α-Myc antibody. Bands were excised and proteins were identified by LC-MS/MS. Identified proteins are marked with an arrow. (B) The Las1-Myc + p426GPD and Las1-Myc + Flag-Grc3 strains were grown in SD-Ura to an OD600 of 0.6. Lysates were then immunoprecipitated with an α-Flag antibody. Immunoprecipitated proteins were separated by SDS-PAGE and analysed by WB using an anti-Flag (Grc3) or an anti-Myc (Las1) antibody. (C) BY4741 + empty vector (p426 GPD), overexpressing Grc3 WT or Grc3 KD were grown and diluted into SD-Ura and OD600 was measured at the indicated time points.
Figure 6.
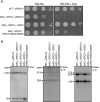
Las1 is required for Grc3 protein stability. (A) The tetO7-GRC3 Las1-Myc strain was grown to an OD600 of 0.1 and treated with (+) or without (−) 10 µg/ml doxycycline for the indicated period of time. Cells were harvested and equal amounts of protein were separated by SDS-PAGE and analysed by WB with an anti-Myc antibody (Las1) and PSTAIRE (loading control). (B) The tetO7-Las1 strain expressing Flag-Grc3 was grown to an OD600 of 0.1 and treated with (+) or without (−) 10 µg/ml doxycycline for the indicated period of time. Cells were harvested and equal amounts of protein were separated by SDS-PAGE and analysed by WB with an anti-Flag antibody (Grc3) and PSTAIRE (loading control). (C) The tetO7-Ipi1 Las1-Myc strain expressing Flag-Grc3 was grown to an OD600 of 0.1 and treated with (+) or without (−) 10 µg/ml doxycycline for the indicated period of time. Cells were harvested and equal amounts of protein were separated by SDS-PAGE and analysed by WB with an anti-Myc antibody (Las1), anti-Flag antibody (Grc3) and PSTAIRE (loading control). (D) Total RNA was harvested from the 2 h time points from the experiments in (A) or (B) (left and right panel, respectively). RT-qPCR analysis was performed to evaluate Las1 or Grc3 mRNA levels as indicated.
Figure 7.
Grc3 kinase activity is required for cell proliferation and pre-rRNA processing. (A) Wild type + pRS413, tetO7-GRC3 + pRS413, tetO7-GRC3 + GRC3, and tetO7-GRC3 + GRC3 K252A/S253A strains were grown in SD-His or SD-His with 10 µg/ml doxycycline and then diluted to and OD600 of 0.05. Ten-fold serial dilutions were performed and 5 µl was plated on SD-His plates with or without 10 µg/ml doxycycline and incubated at 30°C for 48 h. (B) Northern blot analysis of steady-state pre-rRNAs. Wild type + pRS413, tetO7-GRC3 + pRS413, tetO7-GRC3 + GRC3, and tetO7-GRC3 + GRC3 K252A/S253A strains were grown in SD-His with 10 µg/ml doxycycline for 12 h. Equal amounts of RNA were hybridized with probes specific for the indicated region of pre-rRNA. The position of each pre-rRNA is indicated on the left of each panel.
Figure 8.
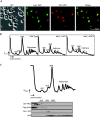
Las1 localizes to the nucleolus and co-fractionates with pre-60S ribosomal particles. (A) Las1-GFP + Sik1-RFP strain was grown in SD-Met to log phase and GFP and RFP signals were analysed by fluorescence microscopy. DAPI was used to costain the nucleus. (B) The tetO7-Parental (WT), tetO7-LAS1 or tetO7-GRC3 strains were grown in YPD to an OD600 of 0.3. Cellular extracts were separated on a 7–47% sucrose gradient and A254 were recorded. The positions of the 40S and 60S pre-ribosomal subunits, 80S ribosomes and polysomes or halfmers are indicated on the profile. (C) The Las1-Myc Rix1-TAP strain expressing Flag-Grc3 was grown in YPD to an OD600 of 0.3. Cellular extracts were separated on a 7–47% sucrose gradient, OD240 were recorded and 600 µl fractions were collected. Proteins were precipitated from each fraction, separated by SDS-PAGE, and analysed by WB with anti-Myc, anti-TAP, anti-Flag or anti-rpL8 antibodies as indicated. The positions of the 40S and 60S pre-ribosomal subunits, 80S ribosomes and polysomes are indicated on the profile.
Figure 9.
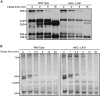
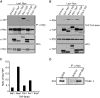
Las1 associates with the Nsa1 and Rix1 pre-60S ribosomal particles. (A) The Las1-Myc, Las1-Myc Ssf1-TAP, Las1-Myc Nsa1-TAP, Las1-Myc Rix1-TAP, Las1-Myc Arx1-TAP or Las1-Myc Enp1-TAP strains were grown to log phase and protein lysates subjected to immunoprecipitation in conditions that preserve the integrity of the pre-ribosomal particles using an anti-Myc antibody. Immunoprecipitated complexes were separated by SDS-PAGE and analysed by WB using an anti-TAP or an anti-Myc (Las1) antibody. WCL, whole cell lysate. (B) The Las1-Myc, Las1-Myc Ssf1-TAP, Las1-Myc Nsa1-TAP, Las1-Myc Rix1-TAP, Las1-Myc Arx1-TAP or Las1-Myc Enp1-TAP strains were grown to log phase and protein lysates subjected to TAP in conditions that preserve the integrity of the pre-ribosomal particles. Purified complexes were separated by SDS-PAGE and analysed by WB using an anti-TAP or an anti-Myc (Las1) antibody. WCL, whole cell lysate. (C) Densitometry quantification of the ratio of Myc-Las1 over each respective TAP-proteins from panel B. (D) The BY4741 and Las1-Myc strains were grown to log phase and protein lysates subjected to immunoprecipitation with an ant-Myc antibody in conditions that preserve the integrity of the pre-ribosomal particles. RNA was extracted from the immuno-purified particles and subjected to northern blot analysis using an E-C2 probe (I). Total RNA was loaded as input.
Similar articles
- Characterization of the molecular crosstalk within the essential Grc3/Las1 pre-rRNA processing complex.
Pillon MC, Sobhany M, Stanley RE. Pillon MC, et al. RNA. 2018 May;24(5):721-738. doi: 10.1261/rna.065037.117. Epub 2018 Feb 9. RNA. 2018. PMID: 29440475 Free PMC article. - Grc3 programs the essential endoribonuclease Las1 for specific RNA cleavage.
Pillon MC, Sobhany M, Borgnia MJ, Williams JG, Stanley RE. Pillon MC, et al. Proc Natl Acad Sci U S A. 2017 Jul 11;114(28):E5530-E5538. doi: 10.1073/pnas.1703133114. Epub 2017 Jun 26. Proc Natl Acad Sci U S A. 2017. PMID: 28652339 Free PMC article. - Nuclease integrated kinase super assemblies (NiKs) and their role in RNA processing.
Pillon MC, Stanley RE. Pillon MC, et al. Curr Genet. 2018 Feb;64(1):183-190. doi: 10.1007/s00294-017-0749-9. Epub 2017 Sep 19. Curr Genet. 2018. PMID: 28929238 Free PMC article. Review. - Role of the yeast ribosomal protein L16 in ribosome biogenesis.
Espinar-Marchena FJ, Fernández-Fernández J, Rodríguez-Galán O, Fernández-Pevida A, Babiano R, de la Cruz J. Espinar-Marchena FJ, et al. FEBS J. 2016 Aug;283(16):2968-85. doi: 10.1111/febs.13797. Epub 2016 Jul 15. FEBS J. 2016. PMID: 27374275 - IT'S 2 for the price of 1: Multifaceted ITS2 processing machines in RNA and DNA maintenance.
Pillon MC, Lo YH, Stanley RE. Pillon MC, et al. DNA Repair (Amst). 2019 Sep;81:102653. doi: 10.1016/j.dnarep.2019.102653. Epub 2019 Jul 8. DNA Repair (Amst). 2019. PMID: 31324529 Free PMC article. Review.
Cited by
- Cryo-EM reveals the architecture of the PELP1-WDR18 molecular scaffold.
Gordon J, Chapus FL, Viverette EG, Williams JG, Deterding LJ, Krahn JM, Borgnia MJ, Rodriguez J, Warren AJ, Stanley RE. Gordon J, et al. Nat Commun. 2022 Nov 9;13(1):6783. doi: 10.1038/s41467-022-34610-0. Nat Commun. 2022. PMID: 36351913 Free PMC article. - Structural overview of macromolecular machines involved in ribosome biogenesis.
Frazier MN, Pillon MC, Kocaman S, Gordon J, Stanley RE. Frazier MN, et al. Curr Opin Struct Biol. 2021 Apr;67:51-60. doi: 10.1016/j.sbi.2020.09.003. Epub 2020 Oct 21. Curr Opin Struct Biol. 2021. PMID: 33099228 Free PMC article. Review. - High resolution landscape of ribosomal RNA processing and surveillance.
An W, Yan Y, Ye K. An W, et al. Nucleic Acids Res. 2024 Sep 23;52(17):10630-10644. doi: 10.1093/nar/gkae606. Nucleic Acids Res. 2024. PMID: 38994562 Free PMC article. - Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Thoms M, Lau B, Cheng J, Fromm L, Denk T, Kellner N, Flemming D, Fischer P, Falquet L, Berninghausen O, Beckmann R, Hurt E. Thoms M, et al. EMBO Rep. 2023 Dec 6;24(12):e57984. doi: 10.15252/embr.202357984. Epub 2023 Nov 3. EMBO Rep. 2023. PMID: 37921038 Free PMC article. - Independent neofunctionalization of Dxo1 in Saccharomyces and Candida led to 25S rRNA processing function.
Hurtig JE, Stuart CJ, van Hoof A. Hurtig JE, et al. RNA. 2024 Nov 18;30(12):1634-1645. doi: 10.1261/rna.080210.124. RNA. 2024. PMID: 39332835
References
- Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. - PubMed
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr. Biol. CB. 2004;14:R1014–R1027. - PubMed
- Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009;21:855–863. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases