Genome-wide methylation profiles reveal quantitative views of human aging rates - PubMed (original) (raw)
. 2013 Jan 24;49(2):359-367.
doi: 10.1016/j.molcel.2012.10.016. Epub 2012 Nov 21.
Justin Guinney # 2, Ling Zhao 3 4 5, Li Zhang 3 4 5 6, Guy Hughes 4 5, SriniVas Sadda 7, Brandy Klotzle 8, Marina Bibikova 8, Jian-Bing Fan 8, Yuan Gao 9, Rob Deconde 1 10, Menzies Chen 1, Indika Rajapakse 11, Stephen Friend 2, Trey Ideker 1 4 10, Kang Zhang 3 4 5
Affiliations
- PMID: 23177740
- PMCID: PMC3780611
- DOI: 10.1016/j.molcel.2012.10.016
Genome-wide methylation profiles reveal quantitative views of human aging rates
Gregory Hannum et al. Mol Cell. 2013.
Abstract
The ability to measure human aging from molecular profiles has practical implications in many fields, including disease prevention and treatment, forensics, and extension of life. Although chronological age has been linked to changes in DNA methylation, the methylome has not yet been used to measure and compare human aging rates. Here, we build a quantitative model of aging using measurements at more than 450,000 CpG markers from the whole blood of 656 human individuals, aged 19 to 101. This model measures the rate at which an individual's methylome ages, which we show is impacted by gender and genetic variants. We also show that differences in aging rates help explain epigenetic drift and are reflected in the transcriptome. Moreover, we show how our aging model is upheld in other human tissues and reveals an advanced aging rate in tumor tissue. Our model highlights specific components of the aging process and provides a quantitative readout for studying the role of methylation in age-related disease.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
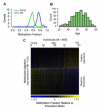
Figure 1. Global data on the aging methylome
(A) A density plot of methylation fraction values for the marker cg16867657, separated by young (green) and old (blue) individuals. (B) A histogram of the age distribution for all individuals. (C) A heat map of the age-associated methylation markers, sorted by the magnitude of association (regression coefficient). The individuals are ordered youngest to oldest. A specific example of an age-associated region is shown in Figure S1. Annotation coincidence tables are also provided as Tables S1, S2.
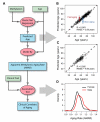
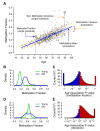
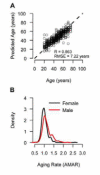
Figure 2. Model predictions and clinical variables
(A) A flow chart of the data (green boxes) and analyses (red ovals) used to generate aging predictions (blue boxes). (B) A comparision of predicted and actual ages for all individuals based on the aging model. (C) Out-of-sample predictions for individuals in the validation cohort. (D) Apparent methylomic aging rate (AMAR) for each individual, based on the aging model without clinical variables. The distribution of aging rates shows faster aging for men than women. A table of the markers used in the aging model are provided as Table S3.
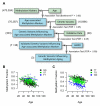
Figure 3. Genetic effects on methylomic aging
(A) We surveyed genomic variants for an association with age-associated methylation markers. 8 genetic variants, corresponding to 14 meQTLs, were chosen for validation. Of these, 7 were significant in the validation cohort and two showed an association with AMAR. (B) A plot of the trend between the methylation marker cg27367526 (STEAP2) and age. The state of variant rs42663 (GTPBP10) causes an offset in this relationship. (C) A second example for cg18404041 and rs2230534 (ITIH1, NEK4). A table of confirmed genetic associations is provided as Table S4.
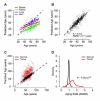
Figure 4. Multi-tissue support
(A) Predictions of age made by the full aging model on the TCGA control samples. There is a high correlation between chronological and predicted age, but each tissue has a different linear intercept and slope. (B) After adjusting the intercept and slope of each tissue, the error of the model is similar to that of the original whole blood data. Age predictions made on cancer samples are presented as Figure S2. (C) Age predictions made on matched normal and tumor samples from TCGA. Predictions are adjusted for the linear offset of the parent tissue (breast, kidney, lung, skin). (D) Tumor samples show a significant increase in AMAR.
Figure 5. Age associations for methylation fraction and deviance
(A) Methylation fraction values for are shown for the marker cg24724428. Over any subset of the cohort, we consider two group methylation statistics: the mean and variance. Marker variance is a measure of the mean methylation deviance, which is defined as the squared difference between an individual’s methylation fraction and their expected methylation fraction. (B) A density plot showing the change in mean methylation with age for the marker cg24724428. Young and old groups are based on the top and bottom 10%. (C) A histogram of the significance of association between the methylation fraction of all markers and age. P-values are signed such that positive values represent an increase of methylation with age. Markers which exceeded the FDR < 0.05 threshold are grouped into the most extreme bins. (D) A density plot showing the change in methylation deviance with age for the marker cg24724428. (E) A histogram in the same form as ‘D’, of the significance of association between the methylation deviance of all markers and age. Aging trends are mapped for CpG islands in Figure S3.
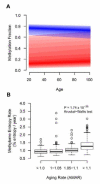
Figure 6. Methylome-wide trends with age
(A) Aggregate regression lines for all methylation markers that increased with age (red) and decreased with age (blue). The darkest color represents the median regression line and the bounds represent the 25% and 75% quantile. Both increasing and decreasing markers trend toward moderate methylation fraction values. (B) An entropy aging rate was calculated as the mean Shannon entropy of age-associated methylation markers divided by chronological age. This was strongly associated with AMAR.
Figure 7. Transcription aging model
(A) We built an aging model using mRNA expression data for genes which showed an aging trend in the methylome. It’s standard error (RMSE = 7.22 years) is increased due to the rounding of ages to the nearest five-year interval in the dataset. (B) Similar to the methylome, the transcriptome shows an increased aging rate for men as compared to women (P < 10−4).
Comment in
- Human epigenetics: Showing your age.
Burgess DJ. Burgess DJ. Nat Rev Genet. 2013 Jan;14(1):6. doi: 10.1038/nrg3391. Epub 2012 Dec 4. Nat Rev Genet. 2013. PMID: 23207909 No abstract available.
Similar articles
- DNA Methylation as a Biomarker of Aging in Epidemiologic Studies.
Lim U, Song MA. Lim U, et al. Methods Mol Biol. 2018;1856:219-231. doi: 10.1007/978-1-4939-8751-1_12. Methods Mol Biol. 2018. PMID: 30178254 Review. - Prediction of biological age and evaluation of genome-wide dynamic methylomic changes throughout human aging.
Amiri Roudbar M, Mousavi SF, Salek Ardestani S, Lopes FB, Momen M, Gianola D, Khatib H. Amiri Roudbar M, et al. G3 (Bethesda). 2021 Jul 14;11(7):jkab112. doi: 10.1093/g3journal/jkab112. G3 (Bethesda). 2021. PMID: 33826720 Free PMC article. - Aging-associated DNA methylation changes in middle-aged individuals: the Young Finns study.
Kananen L, Marttila S, Nevalainen T, Jylhävä J, Mononen N, Kähönen M, Raitakari OT, Lehtimäki T, Hurme M. Kananen L, et al. BMC Genomics. 2016 Feb 9;17:103. doi: 10.1186/s12864-016-2421-z. BMC Genomics. 2016. PMID: 26861258 Free PMC article. - Impacts of Chromatin States and Long-Range Genomic Segments on Aging and DNA Methylation.
Sun D, Yi SV. Sun D, et al. PLoS One. 2015 Jun 19;10(6):e0128517. doi: 10.1371/journal.pone.0128517. eCollection 2015. PLoS One. 2015. PMID: 26091484 Free PMC article. - The role of DNA methylation in epigenetics of aging.
Unnikrishnan A, Freeman WM, Jackson J, Wren JD, Porter H, Richardson A. Unnikrishnan A, et al. Pharmacol Ther. 2019 Mar;195:172-185. doi: 10.1016/j.pharmthera.2018.11.001. Epub 2018 Nov 9. Pharmacol Ther. 2019. PMID: 30419258 Free PMC article. Review.
Cited by
- Understanding the Interplay Between Health Disparities and Epigenomics.
Mancilla VJ, Peeri NC, Silzer T, Basha R, Felini M, Jones HP, Phillips N, Tao MH, Thyagarajan S, Vishwanatha JK. Mancilla VJ, et al. Front Genet. 2020 Aug 20;11:903. doi: 10.3389/fgene.2020.00903. eCollection 2020. Front Genet. 2020. PMID: 32973872 Free PMC article. Review. - Data Mining Suggests That CXCL14 Gene Silencing in Colon Cancer Is Due to Promoter Methylation.
Wang Y, Wang S, Niu Y, Ma B, Li J. Wang Y, et al. Int J Mol Sci. 2023 Nov 7;24(22):16027. doi: 10.3390/ijms242216027. Int J Mol Sci. 2023. PMID: 38003215 Free PMC article. - The mechanism of ageing: primary role of transposable elements in genome disintegration.
Sturm Á, Ivics Z, Vellai T. Sturm Á, et al. Cell Mol Life Sci. 2015 May;72(10):1839-47. doi: 10.1007/s00018-015-1896-0. Epub 2015 Apr 3. Cell Mol Life Sci. 2015. PMID: 25837999 Free PMC article. - DNA methylation study of Huntington's disease and motor progression in patients and in animal models.
Lu AT, Narayan P, Grant MJ, Langfelder P, Wang N, Kwak S, Wilkinson H, Chen RZ, Chen J, Simon Bawden C, Rudiger SR, Ciosi M, Chatzi A, Maxwell A, Hore TA, Aaronson J, Rosinski J, Preiss A, Vogt TF, Coppola G, Monckton D, Snell RG, William Yang X, Horvath S. Lu AT, et al. Nat Commun. 2020 Sep 10;11(1):4529. doi: 10.1038/s41467-020-18255-5. Nat Commun. 2020. PMID: 32913184 Free PMC article. - Epigenetic age acceleration is associated with cardiometabolic risk factors and clinical cardiovascular disease risk scores in African Americans.
Ammous F, Zhao W, Ratliff SM, Mosley TH, Bielak LF, Zhou X, Peyser PA, Kardia SLR, Smith JA. Ammous F, et al. Clin Epigenetics. 2021 Mar 16;13(1):55. doi: 10.1186/s13148-021-01035-3. Clin Epigenetics. 2021. PMID: 33726838 Free PMC article.
References
- Alisch RS, Barwick BG, Chopra P, Myrick LK, Satten GA, Conneely KN, Warren ST. [Accessed May 17, 2012];Age-associated DNA methylation in pediatric populations. Genome Research. 2012 22:623–632. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22300631. - PMC - PubMed
- Atzmon G, Rincon M, Schechter CB, Shuldiner AR, Lipton RB, Bergman A, Barzilai N. [Accessed January 18, 2012];Lipoprotein Genotype and Conserved Pathway for Exceptional Longevity in Humans. PLoS Biol. 2006 4:e113. Available at: UR http://dx.doi.org/10.1371/journal.pbio.0040113,http://dx.doi.org/10.1371.... - DOI - PMC - PubMed
- Barres R, Zierath JR. [Accessed January 18, 2012];DNA methylation in metabolic disorders. The American Journal of Clinical Nutrition. 2011 Available at: http://www.ajcn.org/content/early/2011/02/02/ajcn.110.001933.abstract. - PubMed
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. [Accessed January 31, 2012];DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biology. 2011 12:R10. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21251332. - PMC - PubMed
- Bell JT, Tsai P-C, Yang T-P, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, et al. [Accessed May 17, 2012];Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population. PLoS Genet. 2012 8:e1002629. Available at: http://dx.doi.org/10.1371/journal.pgen.1002629. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- P30 MH062261/MH/NIMH NIH HHS/United States
- P50GM085764,/GM/NIGMS NIH HHS/United States
- R01 EY014428/EY/NEI NIH HHS/United States
- R01E5014811/PHS HHS/United States
- P50 GM085764/GM/NIGMS NIH HHS/United States
- EY021374/EY/NEI NIH HHS/United States
- EY018660/EY/NEI NIH HHS/United States
- EY019270/EY/NEI NIH HHS/United States
- EY014428/EY/NEI NIH HHS/United States
- R01 EY021374/EY/NEI NIH HHS/United States
- R01 ES014811/ES/NIEHS NIH HHS/United States
- R01 EY019270/EY/NEI NIH HHS/United States
- T32 GM007198/GM/NIGMS NIH HHS/United States
- R01 EY018660/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases