MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism - PubMed (original) (raw)
. 2012 Dec;14(12):1336-43.
doi: 10.1038/ncb2622. Epub 2012 Nov 25.
César Cárdenas, Patrick J Doonan, Harish C Chandramoorthy, Krishna M Irrinki, Tünde Golenár, György Csordás, Priyanka Madireddi, Jun Yang, Marioly Müller, Russell Miller, Jill E Kolesar, Jordi Molgó, Brett Kaufman, György Hajnóczky, J Kevin Foskett, Muniswamy Madesh
Affiliations
- PMID: 23178883
- PMCID: PMC3511605
- DOI: 10.1038/ncb2622
MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism
Karthik Mallilankaraman et al. Nat Cell Biol. 2012 Dec.
Erratum in
- Nat Cell Biol. 2013 Jan;15(1):123
- MCUR1 is an essential component of mitochondrial Ca(2+) uptake that regulates cellular metabolism.
Mallilankaraman K, Cárdenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenár T, Csordás G, Madireddi P, Yang J, Müller M, Miller R, Kolesar JE, Molgó J, Kaufman B, Hajnóczky G, Foskett JK, Madesh M. Mallilankaraman K, et al. Nat Cell Biol. 2015 Jul;17(7):953. doi: 10.1038/ncb3202. Nat Cell Biol. 2015. PMID: 26123113 No abstract available.
Abstract
Ca(2+) flux across the mitochondrial inner membrane regulates bioenergetics, cytoplasmic Ca(2+) signals and activation of cell death pathways. Mitochondrial Ca(2+) uptake occurs at regions of close apposition with intracellular Ca(2+) release sites, driven by the inner membrane voltage generated by oxidative phosphorylation and mediated by a Ca(2+) selective ion channel (MiCa; ref. ) called the uniporter whose complete molecular identity remains unknown. Mitochondrial calcium uniporter (MCU) was recently identified as the likely ion-conducting pore. In addition, MICU1 was identified as a mitochondrial regulator of uniporter-mediated Ca(2+) uptake in HeLa cells. Here we identified CCDC90A, hereafter referred to as MCUR1 (mitochondrial calcium uniporter regulator 1), an integral membrane protein required for MCU-dependent mitochondrial Ca(2+) uptake. MCUR1 binds to MCU and regulates ruthenium-red-sensitive MCU-dependent Ca(2+) uptake. MCUR1 knockdown does not alter MCU localization, but abrogates Ca(2+) uptake by energized mitochondria in intact and permeabilized cells. Ablation of MCUR1 disrupts oxidative phosphorylation, lowers cellular ATP and activates AMP kinase-dependent pro-survival autophagy. Thus, MCUR1 is a critical component of a mitochondrial uniporter channel complex required for mitochondrial Ca(2+) uptake and maintenance of normal cellular bioenergetics.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Figures
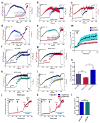
Figure 1. RNAi screen identifies MCUR1 as a regulator of mitochondrial Ca2+ uptake
Changes in 293T cell cytoplasmic (a) and mitochondrial (b) [Ca2+] in response to ionomycin (2.5 μM) were simultaneously measured by fluo-4 and rhod-2 imaging, respectively. Each bar represents one target gene silenced with pooled siRNA. (c) qRT-PCR of MCU, MCUR1 and MICU1 mRNA from mouse tissues (n=3; mean ± s.e.m). (d) qRT-PCR of MCUR1 mRNA from 293T cell clones (n=3; mean ± s.e.m). (e) qRT-PCR of MCUR1 mRNA from HeLa cell clones and of rescued MCUR1 mRNA levels in shHe2 clone (n=3; mean ± s.e.m). The same lentiviral shRNAs were used to generate shHK4 and shHe1 and shHK5 and shHe2, respectively. (f) (Top) MCUR1 protein expression levels and densitometric analysis (n=3; ± s.e.m.). (Bottom) Flag-tagged MCUR1 protein expression in clone shHe2 cells reconstituted with shRNA resistant MCUR1 cDNA plasmid. (g and h) Representative images from movies of HEK 293T NegshRNA or shHK5 cells showing cytosolic (green) and mitochondrial (red) [Ca2+] before (left), during (middle) and after (right) ionomycin exposure. Scale bar: 20 μm. (i–p) Cytoplasmic (green) and mitochondrial matrix (red) [Ca2+] responses in 293T (i–l) and HeLa (m–p) cells challenged with ionomycin or histamine (100 μM), respectively. (n=3) (i) Wild-type 293T cells. (j) Cells expressing negative shRNA. (k) Clone shHK5 (n=4). (l) Quantification of peak rhod-2 fluorescence. **P < 0.01 (mean ± s.e.m.). (m) HeLa cells expressing negative shRNA. (n) Clone shHe2. (o) Clone shHe2 re-expressing MCUR1 (n=3). (p) Quantification of peak rhod-2 fluorescence. *P < 0.05, **P < 0.01 (mean ± s.e.m.). (q) [Ca2+]c and [Ca2+]m signals evoked by ATP (100 μM) and thapsigargin (Tg, 2 μM) were monitored simultaneously using fura2/AM and mtipcam, respectively in control (upper) and MCUR1 KD (middle) HeLa cells. [Ca2+]c calibrated in nM (black), whereas mtipcam fluorescence is inversely normalized to baseline (F0/F) (red). (r) Summary mean [Ca2+]c and [Ca2+]m peaks during ATP stimulation (negShRNA n=29; MCUR1 KD n=36 cells,. *P < 0.05 (mean ± s.e.m.). (s) Increase in bath [Ca2+] (Rfura2) and [Ca2+]m (Rmipcam) signals in response to CaCl2 (1 μM) and IP3 (7.5 μM) addition in permeabilized cells.
Figure 2. MCUR1 is required for Ru360 sensitive mitochondrial Ca2+ uptake but independent of mitochondrial Ca2+ efflux pathway
Digitonin-permeabilized HeLa cells bathed in intracellular-like solution containing thapsigargin (Tg) were loaded with the ΔΨm indicator JC-1 and the Ca2+ indicator Fura2FF, to which pulses of 10 μM Ca2+ were added before addition of mitochondrial uncoupler CCCP (carbonyl cyanide m-chloro phenyl hydrazone). Representative traces from three independent experiments depict simultaneous changes of bath [Ca2+] and ΔΨm in (a) cells expressing negative shRNA, (b) clone shHe1(c) clone shHe2, (d) clone shHe2 re-expressing MCUR1, and (e) HeLa cells stably over-expressing MCUR1. Under similar conditions, 1 μM Ru360 was added before 10 μM Ca2+ pulses until addition of mitochondrial uncoupler CCCP. Representative traces from three independent experiments depict simultaneous changes of bath [Ca2+] and ΔΨm in (f) Negative shRNA cells, (g) MCUR1 knockdown clone shHe1 and (h) shHe2, and (i) in control cells over-expressing MCUR1. Neg shRNA (j) and MCUR1 overexpressing (k) HEK 293T cells were permeabilized with digitonin in intracellular-like medium containing thapsigargin (Tg) and bath [Ca2+] indicator Fura FF, and then pulsed with 10 μM Ca2+. After mitochondrial clearance of bath Ca2+, Ru360 caused elevation of bath [Ca2+], indicating that steady-state bath [Ca2+] after pulse was maintained by balance of MCU-mediated Ca2+ uptake and CGP37157 (10 μM)-sensitive Na+-Ca2+ exchanger-mediated extrusion. CCCP added as indicated. Solid line is mean; shaded areas are ± s.e.m. (n=3). (l) [Ca2+]m efflux rate derived from (j) and (k) during initial 60 s following Ru360 addition (n.s.; not significant, (n=3)). (m) HEK 293T cells stably expressing negative shRNA or MCUR1 shRNA (clone shHe2) and clone ShHe2 re-expressing MCUR1 cells were permeabilized with digitonin in intracellular-like medium containing bath [Ca2+] indicator Fura FF. CCCP added as indicated. Traces show bath [Ca2+] (μM). (solid lines are mean; shaded regions are ± s.e.m. (n= 3). (n) Quantification of total mitochondrial Ca2+ released after CCCP addition. *P < 0.05, **P < 0.01 (mean ± s.e.m).
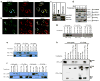
Figure 3. Mitochondrial inner membrane localization and topology of MCUR1 and its interaction with MCU
Confocal images of HeLa cells transiently co-transfected for 48 hrs with (a) GFP-tagged MCUR1 and DsRed Mito plasmids or (b) mRFP-tagged MCUR1 and EYFP-Mito. Scale bar: 20 μm. (c) Immunoblot analysis of Flag-tagged MCUR1 in HeLa cell crude mitochondrial fraction, outer mitochondrial membrane (OMM) and mitoplasts, using antibodies against VDAC1 (OMM protein), COII (integral membrane marker) and Flag. (d) Immunoblot analyses of mitochondria-containing pellet and cytosolic fractions from plasma membrane-permeabilized HeLa cells. Permeabilized cells were treated with or without tBid (50 nM) for outer mitochondrial membrane (OMM) permeabilization and appearance of cytosolic cytochrome c was verified. Intact and OMM permeabilized samples were exposed to Proteinase K for 10 min. These samples were probed using antibodies against HSP60, OXA1, Flag and MCUR1. (e) Reciprocal co-immunoprecipitation of MCU-GFP and MCUR1-Flag transiently expressed in COS7 cells. Representative of four independent experiments. (f) Co-immunoprecipitation of MICU1-Flag with MCU-GFP but not with MCUR1-GFP transiently expressed in COS7 cells. Representative of four independent experiments. (g) Immunoprecipitation of LETM1-GFP with anti-LETM1 failed to pull down MCUR1-Flag in transiently-transfected COS7 cells. (h) Immunoprecipitation with Flag antibody pulled down LETM1-Flag or MCU-Flag transiently expressed in COS7 cells (IP lanes 2 and 3, lower panel), but only co-immunoprecipitated MCUR1-V5 in the MCU-Flag expressing cells (IP lanes 4 vs 5, upper panel), despite lower expression of MCU-Flag in MCUR1-cotransfected cells (compare MCU-Flag and LETM1-Flag expression in MCUR1-cotransfected cells in lysate lanes 4 and 5, bottom panel). Representative of three independent experiments.
Figure 4. MCUR1 is essential for MCU-dependent mitochondrial Ca2+ uptake
(a) qRT-PCR of MCU mRNA from wild type and stable MCUR1 over-expressing HeLa cells that were transiently transfected with scrambled siRNA or siRNA against MCU. ***P < 0.001 (mean ± s.e.m). (b) [Ca2+]m responses to histamine (100 μM) in HeLa cells stably over-expressing MCUR1 and in cells transiently transfected with scrambled siRNA or MCU siRNA, and in stable MCUR1 over-expressing HeLa cells transfected with MCU siRNA. After 48 hr of siRNA transfection, cells were loaded with rhod-2 and [Ca2+]m responses were visualized by confocal microscopy. (solid lines are mean; shaded regions are ± s.e.m.; n= 3). (c) Quantification of peak rhod-2 fluorescence following histamine stimulation. *P < 0.05, ***P < 0.001 (mean ± s.e.m; n=3). (d) qRT-PCR of MCU mRNA from wild type and MCUR1 knockdown HeLa cells that were transiently transfected with MCU cDNA. **P < 0.01, ***P < 0.001 (mean ± s.e.m.; n=3). (e) [Ca2+]m responses to histamine (100μM) in wild-type and MCUR1 (shHe2) knockdown HeLa cells over-expressing MCU. Negative shRNA and MCUR1-shHe2 cells were used as controls. (solid lines are mean; shaded regions are ± s.e.m.; n= 3). (f) Quantification of peak rhod-2 fluorescence following histamine stimulation. ***P < 0.001 (mean ± s.e.m; n=3).
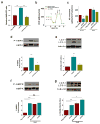
Figure 5. MCUR1 is required for maintenance of cellular bioenergetics
(a) AMP/ATP ratios in stable HeLa cell lines stably expressing negative shRNA, MCUR1 shRNA (clone shHe2) or ShHe2 with MCUR1 re-expressed. **P < 0.01 (mean ± s.e.m.; n=3). (b) O2 consumption rates (OCR) in stable HeLa cells expressing irrelevant shRNA, clone ShHe2, and clone ShHe2 re-expressing MCUR1, exposed sequentially to (a) oligomycin, (b) FCCP, and (c) rotenone plus myzothiazol. (c) Basal and maximal OCR in cells as described in (B). *P < 0.05 (mean ± s.e.m.; n=3). (d) Western blot of phosphorylated and total AMPK (top) and densitometric analysis (bottom) in stable HeLa lines expressing negative shRNA or MCUR1 shRNA (clone sheHe2) and clone shHe2 re-expressing MCUR1. *P < 0.05, **P < 0.01 (mean ± s.e.m.; n=3). (e) Western blot of LC3 or tubulin in stable HeLa lines expressing negative shRNA or MCUR1 shRNA (clone sheHe2) and clone ShHe2 re-expressing MCUR1 (top) and quantification of LC3-II/(LC3-I + LC3-II) (bottom) expressed as fold increase over levels in cells expressing irrelevant shRNA. *P < 0.05, **P < 0.01 (mean ± s.e.m.; n=3). (f and g), as in (d and e). Activation of AMPK (f) and autophagy (**P < 0.001; mean ± s.e.m.; n=3) (g) in absence and presence of InsP3R inhibitor Xestospongin B (XeB). *P < 0.05, **P < 0.001, ns = not significant (mean ± s.e.m.; n=3).
Similar articles
- Mitochondrial Ca2+ uptake 1 (MICU1) and mitochondrial ca2+ uniporter (MCU) contribute to metabolism-secretion coupling in clonal pancreatic β-cells.
Alam MR, Groschner LN, Parichatikanond W, Kuo L, Bondarenko AI, Rost R, Waldeck-Weiermair M, Malli R, Graier WF. Alam MR, et al. J Biol Chem. 2012 Oct 5;287(41):34445-54. doi: 10.1074/jbc.M112.392084. Epub 2012 Aug 17. J Biol Chem. 2012. PMID: 22904319 Free PMC article. - MCUB Regulates the Molecular Composition of the Mitochondrial Calcium Uniporter Channel to Limit Mitochondrial Calcium Overload During Stress.
Lambert JP, Luongo TS, Tomar D, Jadiya P, Gao E, Zhang X, Lucchese AM, Kolmetzky DW, Shah NS, Elrod JW. Lambert JP, et al. Circulation. 2019 Nov 19;140(21):1720-1733. doi: 10.1161/CIRCULATIONAHA.118.037968. Epub 2019 Sep 19. Circulation. 2019. PMID: 31533452 Free PMC article. - SLC25A23 augments mitochondrial Ca²⁺ uptake, interacts with MCU, and induces oxidative stress-mediated cell death.
Hoffman NE, Chandramoorthy HC, Shanmughapriya S, Zhang XQ, Vallem S, Doonan PJ, Malliankaraman K, Guo S, Rajan S, Elrod JW, Koch WJ, Cheung JY, Madesh M. Hoffman NE, et al. Mol Biol Cell. 2014 Mar;25(6):936-47. doi: 10.1091/mbc.E13-08-0502. Epub 2014 Jan 15. Mol Biol Cell. 2014. PMID: 24430870 Free PMC article. - The mitochondrial Ca(2+) uniporter complex.
Foskett JK, Philipson B. Foskett JK, et al. J Mol Cell Cardiol. 2015 Jan;78:3-8. doi: 10.1016/j.yjmcc.2014.11.015. Epub 2014 Nov 22. J Mol Cell Cardiol. 2015. PMID: 25463276 Free PMC article. Review. - From the Identification to the Dissection of the Physiological Role of the Mitochondrial Calcium Uniporter: An Ongoing Story.
Pallafacchina G, Zanin S, Rizzuto R. Pallafacchina G, et al. Biomolecules. 2021 May 23;11(6):786. doi: 10.3390/biom11060786. Biomolecules. 2021. PMID: 34071006 Free PMC article. Review.
Cited by
- The Physiological and Pathological Roles of Mitochondrial Calcium Uptake in Heart.
Lai L, Qiu H. Lai L, et al. Int J Mol Sci. 2020 Oct 17;21(20):7689. doi: 10.3390/ijms21207689. Int J Mol Sci. 2020. PMID: 33080805 Free PMC article. Review. - Cancer cells with defective oxidative phosphorylation require endoplasmic reticulum-to-mitochondria Ca2+ transfer for survival.
Cardenas C, Lovy A, Silva-Pavez E, Urra F, Mizzoni C, Ahumada-Castro U, Bustos G, Jaňa F, Cruz P, Farias P, Mendoza E, Huerta H, Murgas P, Hunter M, Rios M, Cerda O, Georgakoudi I, Zakarian A, Molgó J, Foskett JK. Cardenas C, et al. Sci Signal. 2020 Jul 14;13(640):eaay1212. doi: 10.1126/scisignal.aay1212. Sci Signal. 2020. PMID: 32665411 Free PMC article. - SR-mitochondria communication in adult cardiomyocytes: A close relationship where the Ca2+ has a lot to say.
De la Fuente S, Sheu SS. De la Fuente S, et al. Arch Biochem Biophys. 2019 Mar 15;663:259-268. doi: 10.1016/j.abb.2019.01.026. Epub 2019 Jan 24. Arch Biochem Biophys. 2019. PMID: 30685253 Free PMC article. Review. - EMRE is an essential component of the mitochondrial calcium uniporter complex.
Sancak Y, Markhard AL, Kitami T, Kovács-Bogdán E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, Calvo SE, Goldberger O, Mootha VK. Sancak Y, et al. Science. 2013 Dec 13;342(6164):1379-82. doi: 10.1126/science.1242993. Epub 2013 Nov 14. Science. 2013. PMID: 24231807 Free PMC article. - The Mitochondrial Calcium Uniporter Interacts with Subunit c of the ATP Synthase of Trypanosomes and Humans.
Huang G, Docampo R. Huang G, et al. mBio. 2020 Mar 17;11(2):e00268-20. doi: 10.1128/mBio.00268-20. mBio. 2020. PMID: 32184243 Free PMC article.
References
- Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82:415–424. - PubMed
- Orrenius S, Zhivotovsky B, Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol. 2003;4:552–565. - PubMed
- Denton RM, McCormack JG. The role of calcium in the regulation of mitochondrial metabolism. Biochem Soc Trans. 1980;8:266–268. - PubMed
- Gunter KK, Gunter TE. Transport of calcium by mitochondria. J Bioenerg Biomembr. 1994;26:471–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK051526/DK/NIDDK NIH HHS/United States
- HL086699-01A2S1/HL/NHLBI NIH HHS/United States
- GM56328/GM/NIGMS NIH HHS/United States
- R37 GM056328/GM/NIGMS NIH HHS/United States
- R01 HL119306/HL/NHLBI NIH HHS/United States
- 1S10RR027327-01/RR/NCRR NIH HHS/United States
- R01 MH059937/MH/NIMH NIH HHS/United States
- S10 RR027327/RR/NCRR NIH HHS/United States
- R01 GM056328/GM/NIGMS NIH HHS/United States
- R01 HL086699/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous