Pig domestication and human-mediated dispersal in western Eurasia revealed through ancient DNA and geometric morphometrics - PubMed (original) (raw)
doi: 10.1093/molbev/mss261. Epub 2012 Nov 22.
Linus Girdland Flink, Allowen Evin, Christina Geörg, Bea De Cupere, Wim Van Neer, László Bartosiewicz, Anna Linderholm, Ross Barnett, Joris Peters, Ronny Decorte, Marc Waelkens, Nancy Vanderheyden, François-Xavier Ricaut, Canan Cakirlar, Ozlem Cevik, A Rus Hoelzel, Marjan Mashkour, Azadeh Fatemeh Mohaseb Karimlu, Shiva Sheikhi Seno, Julie Daujat, Fiona Brock, Ron Pinhasi, Hitomi Hongo, Miguel Perez-Enciso, Morten Rasmussen, Laurent Frantz, Hendrik-Jan Megens, Richard Crooijmans, Martien Groenen, Benjamin Arbuckle, Nobert Benecke, Una Strand Vidarsdottir, Joachim Burger, Thomas Cucchi, Keith Dobney, Greger Larson
Affiliations
- PMID: 23180578
- PMCID: PMC3603306
- DOI: 10.1093/molbev/mss261
Pig domestication and human-mediated dispersal in western Eurasia revealed through ancient DNA and geometric morphometrics
Claudio Ottoni et al. Mol Biol Evol. 2013 Apr.
Abstract
Zooarcheological evidence suggests that pigs were domesticated in Southwest Asia ~8,500 BC. They then spread across the Middle and Near East and westward into Europe alongside early agriculturalists. European pigs were either domesticated independently or more likely appeared so as a result of admixture between introduced pigs and European wild boar. As a result, European wild boar mtDNA lineages replaced Near Eastern/Anatolian mtDNA signatures in Europe and subsequently replaced indigenous domestic pig lineages in Anatolia. The specific details of these processes, however, remain unknown. To address questions related to early pig domestication, dispersal, and turnover in the Near East, we analyzed ancient mitochondrial DNA and dental geometric morphometric variation in 393 ancient pig specimens representing 48 archeological sites (from the Pre-Pottery Neolithic to the Medieval period) from Armenia, Cyprus, Georgia, Iran, Syria, and Turkey. Our results reveal the first genetic signatures of early domestic pigs in the Near Eastern Neolithic core zone. We also demonstrate that these early pigs differed genetically from those in western Anatolia that were introduced to Europe during the Neolithic expansion. In addition, we present a significantly more refined chronology for the introduction of European domestic pigs into Asia Minor that took place during the Bronze Age, at least 900 years earlier than previously detected. By the 5th century AD, European signatures completely replaced the endemic lineages possibly coinciding with the widespread demographic and societal changes that occurred during the Anatolian Bronze and Iron Ages.
Figures
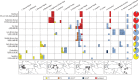
Fig. 1.
A spatiotemporal depiction of ancient pig haplotypes. Rows represent eight chronological periods, and columns pertain to sites organized along a longitudinal axis from west to east. Approximate locations of the archeological sites from which the samples are derived are shown as numbered circles on maps beneath the horizontal axis. Asterisks indicate directly AMS-dated samples. The question mark signifies not enough material was available for AMS dating. Slashed boxes indicate samples on which GMM analyses were performed. Pie charts to the right of each row summarize the haplotype frequencies for each chronological period across all sites. Columns pertain to one or two sites except for two columns that consist of several sites: Armenia (Sevkar-4, Areni-1, Khatunarkh, Shengevit, Lchashen, Tmbatir, Pilorpat, Beniamin, and Tsakaektsi) and Iran (Qaleh Rostam, Qare Doyub, Qelīch Qōīneq, Dasht Qal’eh, Doshan Tepe, Malyan, Mehr Ali, Chogha Gavaneh, and Gohar Tepe).
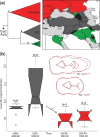
Fig. 2.
Panel (a) depicts a schematic phylogenetic tree derived from an alignment of 267 modern wild boar from western Eurasia. Red, green, and gray triangles refer to the well-supported European, Near Eastern 1 (NE1), and Near Eastern 2 (NE2) clades, respectively. Branches supported by P > 0.99 are indicated by a black circle. A more detailed representation of the tree including support values is presented in
supplementary figure S2_a_
,
Supplementary Material
online. The NE2 clade includes all ancient Near Eastern haplotypes depicted in figure 1. Panel (a) also shows the approximate geographic distribution of modern wild boar belonging to these clades. Areas with overlapping distributions are represented in dark. A more detailed depiction is presented in
supplementary figure S2_b_
,
Supplementary Material
online. Panel (b) presents molar size (M2) and shape (M2 and M3) differences between ancient pigs assigned to European (red) and Near Eastern (gray) mtDNA clades. Differences in shape calculated along linear discriminant analysis (LDA) axes are displayed in overlapping shapes in the upper right. The arrow indicates a statistically significant size reduction in the M2 between European and Near Eastern pigs. Numbers following “_N_=” represent sample sizes, and single and triple asterisks represent significance to the P < 0.05 and P < 0.01 levels.
Similar articles
- Ancient DNA, pig domestication, and the spread of the Neolithic into Europe.
Larson G, Albarella U, Dobney K, Rowley-Conwy P, Schibler J, Tresset A, Vigne JD, Edwards CJ, Schlumbaum A, Dinu A, Balaçsescu A, Dolman G, Tagliacozzo A, Manaseryan N, Miracle P, Van Wijngaarden-Bakker L, Masseti M, Bradley DG, Cooper A. Larson G, et al. Proc Natl Acad Sci U S A. 2007 Sep 25;104(39):15276-81. doi: 10.1073/pnas.0703411104. Epub 2007 Sep 13. Proc Natl Acad Sci U S A. 2007. PMID: 17855556 Free PMC article. - Insular East Asia pig dispersal and vicariance inferred from Asian wild boar genetic evidence.
Li KY, Li KT, Yang CH, Hwang MH, Chang SW, Lin SM, Wu HJ, Basilio EB, Vega RSA, Laude RP, Ju YT. Li KY, et al. J Anim Sci. 2017 Apr;95(4):1451-1466. doi: 10.2527/jas.2016.1117. J Anim Sci. 2017. PMID: 28464072 - Unravelling the complexity of domestication: a case study using morphometrics and ancient DNA analyses of archaeological pigs from Romania.
Evin A, Flink LG, Bălăşescu A, Popovici D, Andreescu R, Bailey D, Mirea P, Lazăr C, Boroneanţ A, Bonsall C, Vidarsdottir US, Brehard S, Tresset A, Cucchi T, Larson G, Dobney K. Evin A, et al. Philos Trans R Soc Lond B Biol Sci. 2015 Jan 19;370(1660):20130616. doi: 10.1098/rstb.2013.0616. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 25487340 Free PMC article. - Mining the pig genome to investigate the domestication process.
Ramos-Onsins SE, Burgos-Paz W, Manunza A, Amills M. Ramos-Onsins SE, et al. Heredity (Edinb). 2014 Dec;113(6):471-84. doi: 10.1038/hdy.2014.68. Epub 2014 Jul 30. Heredity (Edinb). 2014. PMID: 25074569 Free PMC article. Review. - The Sequence Analysis of Mitochondrial DNA Revealed Some Major Centers of Horse Domestications: The Archaeologist's Cut.
Atsenova N, Palova N, Mehandjyiski I, Neov B, Radoslavov G, Hristov P. Atsenova N, et al. J Equine Vet Sci. 2022 Feb;109:103830. doi: 10.1016/j.jevs.2021.103830. Epub 2021 Dec 4. J Equine Vet Sci. 2022. PMID: 34871751 Review.
Cited by
- Swine global genomic resources: insights into wild and domesticated populations.
Benjamin NR, Crooijmans RPMA, Jordan LR, Bolt CR, Schook LB, Schachtschneider KM, Groenen MAM, Roca AL. Benjamin NR, et al. Mamm Genome. 2023 Dec;34(4):520-530. doi: 10.1007/s00335-023-10012-5. Epub 2023 Oct 7. Mamm Genome. 2023. PMID: 37805667 - Anthropogenic hybridization and its influence on the adaptive potential of the Sardinian wild boar (Sus scrofa meridionalis).
Fabbri G, Molinaro L, Mucci N, Pagani L, Scandura M. Fabbri G, et al. J Appl Genet. 2023 Sep;64(3):521-530. doi: 10.1007/s13353-023-00763-x. Epub 2023 Jun 28. J Appl Genet. 2023. PMID: 37369962 Free PMC article. - Distinct traces of mixed ancestry in western commercial pig genomes following gene flow from Chinese indigenous breeds.
Peng Y, Derks MF, Groenen MA, Zhao Y, Bosse M. Peng Y, et al. Front Genet. 2023 Jan 13;13:1070783. doi: 10.3389/fgene.2022.1070783. eCollection 2022. Front Genet. 2023. PMID: 36712875 Free PMC article. - Imputation of Ancient Whole Genome Sus scrofa DNA Introduces Biases Toward Main Population Components in the Reference Panel.
Erven JAM, Çakirlar C, Bradley DG, Raemaekers DCM, Madsen O. Erven JAM, et al. Front Genet. 2022 Jul 12;13:872486. doi: 10.3389/fgene.2022.872486. eCollection 2022. Front Genet. 2022. PMID: 35903348 Free PMC article. - Admixture and breed traceability in European indigenous pig breeds and wild boar using genome-wide SNP data.
Dadousis C, Muñoz M, Óvilo C, Fabbri MC, Araújo JP, Bovo S, Potokar MČ, Charneca R, Crovetti A, Gallo M, García-Casco JM, Karolyi D, Kušec G, Martins JM, Mercat MJ, Pugliese C, Quintanilla R, Radović Č, Razmaite V, Ribani A, Riquet J, Savić R, Schiavo G, Škrlep M, Tinarelli S, Usai G, Zimmer C, Fontanesi L, Bozzi R. Dadousis C, et al. Sci Rep. 2022 May 5;12(1):7346. doi: 10.1038/s41598-022-10698-8. Sci Rep. 2022. PMID: 35513520 Free PMC article.
References
- Bocquet-Appel JP. When the world’s population took off: the springboard of the Neolithic Demographic Transition. Science. 2011;333:560–561. - PubMed
- Bramanti B, Thomas MG, Haak W, et al. (16 co-authors) Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326:137–140. - PubMed
- Brock F, Higham TFG, Ditchfield P, Bronk Ramsey C. Current pretreatment methods for AMS radiocarbon dating at the Oxford Radiocarbon Accelerator Unit (ORAU) Radiocarbon. 2010;52:103–112.
- Bryce T. The kingdom of the Hittites. Oxford: Oxford University Press; 2005.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials