Decoding human cytomegalovirus - PubMed (original) (raw)
. 2012 Nov 23;338(6110):1088-93.
doi: 10.1126/science.1227919.
Ben Weisburd, Annette Michalski, Vu Thuy Khanh Le, Marco Y Hein, Sheng-Xiong Huang, Ming Ma, Ben Shen, Shu-Bing Qian, Hartmut Hengel, Matthias Mann, Nicholas T Ingolia, Jonathan S Weissman
Affiliations
- PMID: 23180859
- PMCID: PMC3817102
- DOI: 10.1126/science.1227919
Decoding human cytomegalovirus
Noam Stern-Ginossar et al. Science. 2012.
Abstract
The human cytomegalovirus (HCMV) genome was sequenced 20 years ago. However, like those of other complex viruses, our understanding of its protein coding potential is far from complete. We used ribosome profiling and transcript analysis to experimentally define the HCMV translation products and follow their temporal expression. We identified hundreds of previously unidentified open reading frames and confirmed a fraction by means of mass spectrometry. We found that regulated use of alternative transcript start sites plays a broad role in enabling tight temporal control of HCMV protein expression and allowing multiple distinct polypeptides to be generated from a single genomic locus. Our results reveal an unanticipated complexity to the HCMV coding capacity and illustrate the role of regulated changes in transcript start sites in generating this complexity.
Figures
Fig. 1
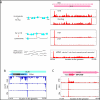
Ribosome profiling of HCMV infected cells. (A) Ribosome occupancies following various treatments (illustrated on the left); cycloheximide (CHX), No-drug, harringtonine (Harr) and LTM together with mRNA profiles of the UL25 gene at 72 hpi. An arrow marks the mRNA start. (B,C) Ribosome occupancy profiles for UL38 (B) and UL10(C) genes that contain internal initiations. The grey area symbolizes a low complexity region.
Fig. 2
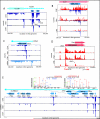
Many ribosome footprints do not correspond to previously annotated ORFs. (A) Ribosome occupancy profiles for the leader region of UL139 gene. (B) Ribosome occupancy profiles of plus and minus strands (red and blue respectively) for the UL91 gene. (C) mRNA and ribosome occupancy profiles for a novel short ORF. (D) Ribosome occupancies around a short ORF that initiates at a CUG codon. (E) Ribosome occupancy profiles for RNA β2.7. The upper panels show the annotated MS/MS spectra of two unique peptides originating from ORFL6C and ORFL7C.
Fig. 3
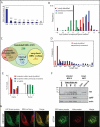
Annotating the HCMV translated ORFs. (A) Fold enrichment of AUG and near-cognate codons at predicted sites of translation initiation compared to their genomic distribution. (B) The ribosome footprints occupancy after LTM treatment at each start codon (relative to the median density across the gene) is depicted for the previously annotated ORFs (blue) and newly identified ORFs (red and empty red for ORFs that were removed). The occupancy at a codon five positions downstream of the start codon is depicted as a control (green). (C) Venn diagram summarizing the HCMV translated ORFs. Note 53 ORFs were initially identified by manual inspection (see text). (D) The lengths distribution of newly identified ORFs (red) and previously annotated ORFs (blue). (E) Position of 30-nt ribosome footprints relative to the reading frame in the newly identified ORFs (red) and previously annotated ORFs (blue). (F) MRC-5 cells were mock-treated or infected with TB40-US33A-HA and protein lysates were analyzed by western blotting with indicated antibodies. (G) HeLa cells were transfected with GFP fusion proteins together with an ER marker (KDEL-mCherry) or stained with MitoTracker Red and imaged by confocal microscopy.
Fig. 4
A major source of ORFs diversity during infection originates from alternative transcripts starts. (A) The mRNA and ribosome occupancy profiles around US18-US20 loci at different infection times (marked on the left). Small arrows denote the different mRNA starts and the corresponding mRNAs are illustrated (upper part). The lower panel shows an expanded view of the US18 locus at 72 hpi and includes the harringtonine and LTM profiles (the internal initiation is marked with a star). (B) Total RNA extracted at different time points during infection was subjected to Northern blotting for ORFS346C.1
Similar articles
- A cluster of 3' coterminal transcripts from US12-US17 locus of human cytomegalovirus.
Lu Y, Ma Y, Liu Z, Han L, Gao S, Zheng B, Liu C, Qi Y, Sun Z, Huang Y, Ruan Q. Lu Y, et al. Virus Genes. 2016 Jun;52(3):334-45. doi: 10.1007/s11262-016-1308-z. Epub 2016 Mar 1. Virus Genes. 2016. PMID: 26931512 - Contributions of the Human Cytomegalovirus UL30-Associated Open Reading Frames to Infection.
Monaghan M, Munger J. Monaghan M, et al. J Virol. 2021 Apr 12;95(9):e02417-20. doi: 10.1128/JVI.02417-20. Print 2021 Apr 12. J Virol. 2021. PMID: 33568511 Free PMC article. - Sequence variability of human cytomegalovirus UL146 and UL147 genes in low-passage clinical isolates.
He R, Ruan Q, Qi Y, Ma YP, Huang YJ, Sun ZR, Ji YH. He R, et al. Intervirology. 2006;49(4):215-23. doi: 10.1159/000091468. Epub 2006 Feb 16. Intervirology. 2006. PMID: 16491016 - Decoding viral infection by ribosome profiling.
Stern-Ginossar N. Stern-Ginossar N. J Virol. 2015 Jun;89(12):6164-6. doi: 10.1128/JVI.02528-14. Epub 2015 Mar 25. J Virol. 2015. PMID: 25810547 Free PMC article. Review. - Human cytomegalovirus genome.
Murphy E, Shenk T. Murphy E, et al. Curr Top Microbiol Immunol. 2008;325:1-19. doi: 10.1007/978-3-540-77349-8_1. Curr Top Microbiol Immunol. 2008. PMID: 18637497 Review.
Cited by
- Targeted Mutagenesis of Guinea Pig Cytomegalovirus Using CRISPR/Cas9-Mediated Gene Editing.
Bierle CJ, Anderholm KM, Wang JB, McVoy MA, Schleiss MR. Bierle CJ, et al. J Virol. 2016 Jul 11;90(15):6989-6998. doi: 10.1128/JVI.00139-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27226370 Free PMC article. - Characterizing inactive ribosomes in translational profiling.
Liu B, Qian SB. Liu B, et al. Translation (Austin). 2016 Jan 8;4(1):e1138018. doi: 10.1080/21690731.2015.1138018. eCollection 2016 Jan-Jun. Translation (Austin). 2016. PMID: 27335722 Free PMC article. - Identification of a herpes simplex virus 1 gene encoding neurovirulence factor by chemical proteomics.
Kato A, Adachi S, Kawano S, Takeshima K, Watanabe M, Kitazume S, Sato R, Kusano H, Koyanagi N, Maruzuru Y, Arii J, Hatta T, Natsume T, Kawaguchi Y. Kato A, et al. Nat Commun. 2020 Sep 29;11(1):4894. doi: 10.1038/s41467-020-18718-9. Nat Commun. 2020. PMID: 32994400 Free PMC article. - Analysis and mapping of a 3' coterminal transcription unit derived from human cytomegalovirus open reading frames UL30-UL32.
Ma Y, Gao S, Wang L, Wang N, Li M, Zheng B, Qi Y, Sun Z, Liu W, Ruan Q. Ma Y, et al. Virol J. 2013 Feb 27;10:65. doi: 10.1186/1743-422X-10-65. Virol J. 2013. PMID: 23446136 Free PMC article. - The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction.
Turner DL, Mathias RA. Turner DL, et al. Front Cell Dev Biol. 2022 Nov 25;10:1053139. doi: 10.3389/fcell.2022.1053139. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506089 Free PMC article. Review.
References
- Mocarski ES, Shenk T, Pass RF. Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 2007.
- Davison AJ, et al. J Gen Virol. 2003;84:17. - PubMed
- Murphy E, Shenk T. Curr Top Microbiol Immunol. 2008;325:1. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases