NCBI GEO: archive for functional genomics data sets--update - PubMed (original) (raw)
. 2013 Jan;41(Database issue):D991-5.
doi: 10.1093/nar/gks1193. Epub 2012 Nov 27.
Stephen E Wilhite, Pierre Ledoux, Carlos Evangelista, Irene F Kim, Maxim Tomashevsky, Kimberly A Marshall, Katherine H Phillippy, Patti M Sherman, Michelle Holko, Andrey Yefanov, Hyeseung Lee, Naigong Zhang, Cynthia L Robertson, Nadezhda Serova, Sean Davis, Alexandra Soboleva
Affiliations
- PMID: 23193258
- PMCID: PMC3531084
- DOI: 10.1093/nar/gks1193
NCBI GEO: archive for functional genomics data sets--update
Tanya Barrett et al. Nucleic Acids Res. 2013 Jan.
Abstract
The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) is an international public repository for high-throughput microarray and next-generation sequence functional genomic data sets submitted by the research community. The resource supports archiving of raw data, processed data and metadata which are indexed, cross-linked and searchable. All data are freely available for download in a variety of formats. GEO also provides several web-based tools and strategies to assist users to query, analyse and visualize data. This article reports current status and recent database developments, including the release of GEO2R, an R-based web application that helps users analyse GEO data.
Figures
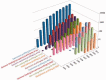
Figure 1.
Distribution of the number and types of selected studies released by GEO each year since inception. Users can explore and download historical submission numbers using the ‘history’ page at
http://www.ncbi.nlm.nih.gov/geo/summary/?type=history
, as well as constructing GEO DataSet database queries for specific data types and date ranges using the ‘DataSet type’ and ‘publication date’ fields as described at
http://www.ncbi.nlm.nih.gov/geo/info/qqtutorial.html
.
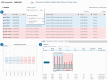
Figure 2.
GEO2R screenshots. After selecting ‘analyse with GEO2R’ on series record GSE18388 (19), the user is presented with a table of the samples in that study and their descriptions (Panel 1). In this case, two sample groups are defined, and four samples are assigned to each group. The user can view the distribution of the sample values using the boxplot feature (Panel 2) and click the ‘Top250’ button to retrieve a table of the top 250 differentially expressed genes with statistics and gene annotation (Panel 3). The top hit is clicked to reveal the expression profile chart for that gene.
Similar articles
- NCBI GEO: archive for gene expression and epigenomics data sets: 23-year update.
Clough E, Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Lee H, Zhang N, Serova N, Wagner L, Zalunin V, Kochergin A, Soboleva A. Clough E, et al. Nucleic Acids Res. 2024 Jan 5;52(D1):D138-D144. doi: 10.1093/nar/gkad965. Nucleic Acids Res. 2024. PMID: 37933855 Free PMC article. - NCBI GEO: archive for functional genomics data sets--10 years on.
Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A. Barrett T, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D1005-10. doi: 10.1093/nar/gkq1184. Epub 2010 Nov 21. Nucleic Acids Res. 2011. PMID: 21097893 Free PMC article. - NCBI GEO: archive for high-throughput functional genomic data.
Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R. Barrett T, et al. Nucleic Acids Res. 2009 Jan;37(Database issue):D885-90. doi: 10.1093/nar/gkn764. Epub 2008 Oct 21. Nucleic Acids Res. 2009. PMID: 18940857 Free PMC article. - Gene expression omnibus: microarray data storage, submission, retrieval, and analysis.
Barrett T, Edgar R. Barrett T, et al. Methods Enzymol. 2006;411:352-69. doi: 10.1016/S0076-6879(06)11019-8. Methods Enzymol. 2006. PMID: 16939800 Free PMC article. Review. - Reuse of public genome-wide gene expression data.
Rung J, Brazma A. Rung J, et al. Nat Rev Genet. 2013 Feb;14(2):89-99. doi: 10.1038/nrg3394. Epub 2012 Dec 27. Nat Rev Genet. 2013. PMID: 23269463 Review.
Cited by
- Targeting the tumor stroma: integrative analysis reveal GATA2 and TORYAIP1 as novel prognostic targets in breast and ovarian cancer.
Erceylan ÖF, Savaş A, Göv E. Erceylan ÖF, et al. Turk J Biol. 2021 Apr 20;45(2):127-137. doi: 10.3906/biy-2010-39. eCollection 2021. Turk J Biol. 2021. PMID: 33907495 Free PMC article. - A dominant role for the methyl-CpG-binding protein Mbd2 in controlling Th2 induction by dendritic cells.
Cook PC, Owen H, Deaton AM, Borger JG, Brown SL, Clouaire T, Jones GR, Jones LH, Lundie RJ, Marley AK, Morrison VL, Phythian-Adams AT, Wachter E, Webb LM, Sutherland TE, Thomas GD, Grainger JR, Selfridge J, McKenzie AN, Allen JE, Fagerholm SC, Maizels RM, Ivens AC, Bird A, MacDonald AS. Cook PC, et al. Nat Commun. 2015 Apr 24;6:6920. doi: 10.1038/ncomms7920. Nat Commun. 2015. PMID: 25908537 Free PMC article. - Fructose-1,6-bisphosphatase aggravates oxidative stress-induced apoptosis in asthma by suppressing the Nrf2 pathway.
Hu J, Wang J, Li C, Shang Y. Hu J, et al. J Cell Mol Med. 2021 Jun;25(11):5001-5014. doi: 10.1111/jcmm.16439. Epub 2021 May 7. J Cell Mol Med. 2021. PMID: 33960626 Free PMC article. - Unveiling prognostics biomarkers of tyrosine metabolism reprogramming in liver cancer by cross-platform gene expression analyses.
Nguyen TN, Nguyen HQ, Le DH. Nguyen TN, et al. PLoS One. 2020 Jun 15;15(6):e0229276. doi: 10.1371/journal.pone.0229276. eCollection 2020. PLoS One. 2020. PMID: 32542016 Free PMC article. - Publisher’s Note:Abstraction for data integration:Fusing mammalian molecular, cellular and phenotype big datasets for better knowledge extraction.
Rouillard AD, Wang Z, Ma'ayan A. Rouillard AD, et al. Comput Biol Chem. 2015 Oct;58:104-19. doi: 10.1016/j.compbiolchem.2015.06.003. Comput Biol Chem. 2015. PMID: 26101093 Free PMC article.
References
- Microarray standards at last. Nature. 2002;419:323. - PubMed
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources