Clone DB: an integrated NCBI resource for clone-associated data - PubMed (original) (raw)
. 2013 Jan;41(Database issue):D1070-8.
doi: 10.1093/nar/gks1164. Epub 2012 Nov 27.
Affiliations
- PMID: 23193260
- PMCID: PMC3531087
- DOI: 10.1093/nar/gks1164
Clone DB: an integrated NCBI resource for clone-associated data
Valerie A Schneider et al. Nucleic Acids Res. 2013 Jan.
Abstract
The National Center for Biotechnology Information (NCBI) Clone DB (http://www.ncbi.nlm.nih.gov/clone/) is an integrated resource providing information about and facilitating access to clones, which serve as valuable research reagents in many fields, including genome sequencing and variation analysis. Clone DB represents an expansion and replacement of the former NCBI Clone Registry and has records for genomic and cell-based libraries and clones representing more than 100 different eukaryotic taxa. Records provide details of library construction, associated sequences, map positions and information about resource distribution. Clone DB is indexed in the NCBI Entrez system and can be queried by fields that include organism, clone name, gene name and sequence identifier. Whenever possible, genomic clones are mapped to reference assemblies and their map positions provided in clone records. Clones mapping to specific genomic regions can also be searched for using the NCBI Clone Finder tool, which accepts queries based on sequence coordinates or features such as gene or transcript names. Clone DB makes reports of library, clone and placement data on its FTP site available for download. With Clone DB, users now have available to them a centralized resource that provides them with the tools they will need to make use of these important research reagents.
Figures
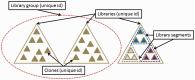
Figure 1.
Schematic illustrating data relationships in Clone DB. Three different libraries (large open triangles) are represented. Libraries sharing a common set of user-specified attributes can be assigned to a library group (dashed circle). Within Clone DB, every clone (small filled triangles) must be associated with an existing library record. In instances where subsets of clones belonging to the same library exhibit different attributes (small triangles at far right with different fill colors), library segments are used to distinguish clones sharing common attributes (dashed open triangles). Library groups, libraries and clones are all assigned unique numeric identifiers within Clone DB; library segments are attributes of library records.
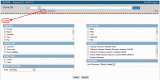
Figure 2.
Limits facilitate searches of Clone DB. Top: screen shot of the header found on all Clone DB web pages. Complex text-based queries can be directly entered into the search box. Beneath the box, the ‘Limits’ and ‘Advanced’ options provide links to other pages that provide alternate approaches to querying the database. Bottom (red arrow): screen shot of ‘Limits’ page for Clone DB. Users may restrict their searches by selecting from a pre-defined set of commonly used Clone DB search terms for several indexed fields. To further refine searches, users may also combine selected limits with queries entered into the text box.
Figure 3.
Genomic clone placement displays in Clone DB. Left panel: screen shot taken from a genomic clone record display (
http://www.ncbi.nlm.nih.gov/clone/5429/
). The ‘Genome View’ tab contains a graphic representation of clone placements in the context of assembly features via the NCBI Sequence Viewer. Blue/tan rectangles: assembly components. Green bars: NCBI genes, with RefSeq transcripts and proteins immediately below. At this zoom level, clone placements appear as a series of horizontal lines with triangular ends. The selected placement for the clone of record appears first and is highlighted with a gray box. Additional placements represent other clones from the same genomic library. The black arrow points to the pop-up menu for the selected placement. Right panel: legend for genomic clone placement displays in NCBI Sequence Viewer (legend also found at:
http://www.ncbi.nlm.nih.gov/clone/content/placements/
).
Figure 4.
Clone Finder search results. Top: search interface. Users can query a selected assembly by coordinate or feature (circle). A series of filters (red box) can be applied to restrict search results. This example shows a search for clones belonging to selected libraries that are mapped to the genomic interval containing NCAM1 in the GRCh37.p5 human reference assembly. Bottom: search results. The ideogram at top provides a chromosomal context for the search (red arrow). The graphic display below shows the chromosome coordinates and sequence identifiers of the contigs and components that comprise it, along with annotated RefSeq and Ensembl genes and transcripts (black arrows). Beneath this, placements from each library are presented in separated panels. As noted, concordant placements for clones within the region are colored in green and discordant placements in red. The display of these placements, like the option to the display results in tabular format, can be toggled on and off (red rectangle). Search results can also be downloaded to Excel for further analysis (red circle).
Similar articles
- Gene: a gene-centered information resource at NCBI.
Brown GR, Hem V, Katz KS, Ovetsky M, Wallin C, Ermolaeva O, Tolstoy I, Tatusova T, Pruitt KD, Maglott DR, Murphy TD. Brown GR, et al. Nucleic Acids Res. 2015 Jan;43(Database issue):D36-42. doi: 10.1093/nar/gku1055. Epub 2014 Oct 29. Nucleic Acids Res. 2015. PMID: 25355515 Free PMC article. - Entrez Gene: gene-centered information at NCBI.
Maglott D, Ostell J, Pruitt KD, Tatusova T. Maglott D, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D52-7. doi: 10.1093/nar/gkq1237. Epub 2010 Nov 28. Nucleic Acids Res. 2011. PMID: 21115458 Free PMC article. - Entrez Gene: gene-centered information at NCBI.
Maglott D, Ostell J, Pruitt KD, Tatusova T. Maglott D, et al. Nucleic Acids Res. 2005 Jan 1;33(Database issue):D54-8. doi: 10.1093/nar/gki031. Nucleic Acids Res. 2005. PMID: 15608257 Free PMC article. Updated. - Education resources of the National Center for Biotechnology Information.
Cooper PS, Lipshultz D, Matten WT, McGinnis SD, Pechous S, Romiti ML, Tao T, Valjavec-Gratian M, Sayers EW. Cooper PS, et al. Brief Bioinform. 2010 Nov;11(6):563-9. doi: 10.1093/bib/bbq022. Epub 2010 Jun 22. Brief Bioinform. 2010. PMID: 20570844 Free PMC article. Review. - Integration of cytogenetic data with genome maps and available probes: present status and future promise.
Kirsch IR, Ried T. Kirsch IR, et al. Semin Hematol. 2000 Oct;37(4):420-8. doi: 10.1016/s0037-1963(00)90021-0. Semin Hematol. 2000. PMID: 11071363 Review.
Cited by
- A New Chicken Genome Assembly Provides Insight into Avian Genome Structure.
Warren WC, Hillier LW, Tomlinson C, Minx P, Kremitzki M, Graves T, Markovic C, Bouk N, Pruitt KD, Thibaud-Nissen F, Schneider V, Mansour TA, Brown CT, Zimin A, Hawken R, Abrahamsen M, Pyrkosz AB, Morisson M, Fillon V, Vignal A, Chow W, Howe K, Fulton JE, Miller MM, Lovell P, Mello CV, Wirthlin M, Mason AS, Kuo R, Burt DW, Dodgson JB, Cheng HH. Warren WC, et al. G3 (Bethesda). 2017 Jan 5;7(1):109-117. doi: 10.1534/g3.116.035923. G3 (Bethesda). 2017. PMID: 27852011 Free PMC article. - Single haplotype assembly of the human genome from a hydatidiform mole.
Steinberg KM, Schneider VA, Graves-Lindsay TA, Fulton RS, Agarwala R, Huddleston J, Shiryev SA, Morgulis A, Surti U, Warren WC, Church DM, Eichler EE, Wilson RK. Steinberg KM, et al. Genome Res. 2014 Dec;24(12):2066-76. doi: 10.1101/gr.180893.114. Epub 2014 Nov 4. Genome Res. 2014. PMID: 25373144 Free PMC article. - Database resources of the National Center for Biotechnology Information.
NCBI Resource Coordinators. NCBI Resource Coordinators. Nucleic Acids Res. 2013 Jan;41(Database issue):D8-D20. doi: 10.1093/nar/gks1189. Epub 2012 Nov 27. Nucleic Acids Res. 2013. PMID: 23193264 Free PMC article. - Database resources of the National Center for Biotechnology Information.
NCBI Resource Coordinators. NCBI Resource Coordinators. Nucleic Acids Res. 2015 Jan;43(Database issue):D6-17. doi: 10.1093/nar/gku1130. Epub 2014 Nov 14. Nucleic Acids Res. 2015. PMID: 25398906 Free PMC article. - An integrated chromosome-scale genome assembly of the Masai giraffe (Giraffa camelopardalis tippelskirchi).
Farré M, Li Q, Darolti I, Zhou Y, Damas J, Proskuryakova AA, Kulemzina AI, Chemnick LG, Kim J, Ryder OA, Ma J, Graphodatsky AS, Zhang G, Larkin DM, Lewin HA. Farré M, et al. Gigascience. 2019 Aug 1;8(8):giz090. doi: 10.1093/gigascience/giz090. Gigascience. 2019. PMID: 31367745 Free PMC article.
References
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous