KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters - PubMed (original) (raw)
. 2013 Jan;41(Database issue):D353-7.
doi: 10.1093/nar/gks1239. Epub 2012 Nov 27.
Toshiaki Katayama, Masumi Itoh, Kazushi Hiranuka, Shuichi Kawashima, Yuki Moriya, Shujiro Okuda, Michihiro Tanaka, Toshiaki Tokimatsu, Yoshihiro Yamanishi, Akiyasu C Yoshizawa, Minoru Kanehisa, Susumu Goto
Affiliations
- PMID: 23193276
- PMCID: PMC3531156
- DOI: 10.1093/nar/gks1239
KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters
Akihiro Nakaya et al. Nucleic Acids Res. 2013 Jan.
Abstract
The identification of orthologous genes in an increasing number of fully sequenced genomes is a challenging issue in recent genome science. Here we present KEGG OC (http://www.genome.jp/tools/oc/), a novel database of ortholog clusters (OCs). The current version of KEGG OC contains 1 176 030 OCs, obtained by clustering 8 357 175 genes in 2112 complete genomes (153 eukaryotes, 1830 bacteria and 129 archaea). The OCs were constructed by applying the quasi-clique-based clustering method to all possible protein coding genes in all complete genomes, based on their amino acid sequence similarities. It is computationally efficient to calculate OCs, which enables to regularly update the contents. KEGG OC has the following two features: (i) It consists of all complete genomes of a wide variety of organisms from three domains of life, and the number of organisms is the largest among the existing databases; and (ii) It is compatible with the KEGG database by sharing the same sets of genes and identifiers, which leads to seamless integration of OCs with useful components in KEGG such as biological pathways, pathway modules, functional hierarchy, diseases and drugs. The KEGG OC resources are accessible via OC Viewer that provides an interactive visualization of OCs at different taxonomic levels.
Figures
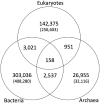
Figure 1.
Distribution of OCs in KEGG OC across three domains: eukaryotes, bacteria and archaea. The number indicates the number of OCs consisting of multiple genes, whereas the number in parenthesis indicates the number of singletons (OCs consisting of a single gene).
Figure 2.
An example of the output page of OC Viewer of query ‘eco:b0002’ (an example of KEGG GENES ID for a gene of E. coli K-12 MG1655) as an input. The PC column shows the PCs (eco.14, ecj.17, ecd.113, etc.). These PCs are aggregated into a TC named Escherichia_col.10890 at the higher taxonomic level indicated in 5th column. As the aggregation of the TCs is iterated from the 5th column to the 2nd column in the OC table, these PCs are merged to the top-level cluster OC.149602. By using the slider at the bottom left, one can focus to arbitrary depth in the taxonomic tree indicated at the bottom right.
Similar articles
- Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea.
Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Makarova KS, et al. Biol Direct. 2007 Nov 27;2:33. doi: 10.1186/1745-6150-2-33. Biol Direct. 2007. PMID: 18042280 Free PMC article. - Automatic detection of conserved gene clusters in multiple genomes by graph comparison and P-quasi grouping.
Fujibuchi W, Ogata H, Matsuda H, Kanehisa M. Fujibuchi W, et al. Nucleic Acids Res. 2000 Oct 15;28(20):4029-36. doi: 10.1093/nar/28.20.4029. Nucleic Acids Res. 2000. PMID: 11024184 Free PMC article. - KEGG for taxonomy-based analysis of pathways and genomes.
Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. Kanehisa M, et al. Nucleic Acids Res. 2023 Jan 6;51(D1):D587-D592. doi: 10.1093/nar/gkac963. Nucleic Acids Res. 2023. PMID: 36300620 Free PMC article. - Comparative Genomics for Prokaryotes.
Setubal JC, Almeida NF, Wattam AR. Setubal JC, et al. Methods Mol Biol. 2018;1704:55-78. doi: 10.1007/978-1-4939-7463-4_3. Methods Mol Biol. 2018. PMID: 29277863 Review. - Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world.
Koonin EV, Wolf YI. Koonin EV, et al. Nucleic Acids Res. 2008 Dec;36(21):6688-719. doi: 10.1093/nar/gkn668. Epub 2008 Oct 23. Nucleic Acids Res. 2008. PMID: 18948295 Free PMC article. Review.
Cited by
- Metagenome survey of a multispecies and alga-associated biofilm revealed key elements of bacterial-algal interactions in photobioreactors.
Krohn-Molt I, Wemheuer B, Alawi M, Poehlein A, Güllert S, Schmeisser C, Pommerening-Röser A, Grundhoff A, Daniel R, Hanelt D, Streit WR. Krohn-Molt I, et al. Appl Environ Microbiol. 2013 Oct;79(20):6196-206. doi: 10.1128/AEM.01641-13. Epub 2013 Aug 2. Appl Environ Microbiol. 2013. PMID: 23913425 Free PMC article. - Transcription factor and microRNA-regulated network motifs for cancer and signal transduction networks.
Hsieh WT, Tzeng KR, Ciou JS, Tsai JJ, Kurubanjerdjit N, Huang CH, Ng KL. Hsieh WT, et al. BMC Syst Biol. 2015;9 Suppl 1(Suppl 1):S5. doi: 10.1186/1752-0509-9-S1-S5. Epub 2015 Jan 21. BMC Syst Biol. 2015. PMID: 25707690 Free PMC article. - Proposal for a new therapy for drug-resistant malaria using Plasmodium synthetic lethality inference.
Lee SJ, Seo E, Cho Y. Lee SJ, et al. Int J Parasitol Drugs Drug Resist. 2013 Jun 28;3:119-28. doi: 10.1016/j.ijpddr.2013.06.001. eCollection 2013 Dec. Int J Parasitol Drugs Drug Resist. 2013. PMID: 24533301 Free PMC article. - KCF-S: KEGG Chemical Function and Substructure for improved interpretability and prediction in chemical bioinformatics.
Kotera M, Tabei Y, Yamanishi Y, Moriya Y, Tokimatsu T, Kanehisa M, Goto S. Kotera M, et al. BMC Syst Biol. 2013;7 Suppl 6(Suppl 6):S2. doi: 10.1186/1752-0509-7-S6-S2. Epub 2013 Dec 13. BMC Syst Biol. 2013. PMID: 24564846 Free PMC article. - Oncofinder, a new method for the analysis of intracellular signaling pathway activation using transcriptomic data.
Buzdin AA, Zhavoronkov AA, Korzinkin MB, Venkova LS, Zenin AA, Smirnov PY, Borisov NM. Buzdin AA, et al. Front Genet. 2014 Mar 25;5:55. doi: 10.3389/fgene.2014.00055. eCollection 2014. Front Genet. 2014. PMID: 24723936 Free PMC article.
References
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. - PubMed