Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria - PubMed (original) (raw)
Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria
Rajatava Basu et al. Immunity. 2012.
Abstract
Interleukin-22 (IL-22) is central to host protection against bacterial infections at barrier sites. Both innate lymphoid cells (ILCs) and T cells produce IL-22. However, the specific contributions of CD4(+) T cells and their developmental origins are unclear. We found that the enteric pathogen Citrobacter rodentium induced sequential waves of IL-22-producing ILCs and CD4(+) T cells that were each critical to host defense during a primary infection. Whereas IL-22 production by ILCs was strictly IL-23 dependent, development of IL-22-producing CD4(+) T cells occurred via an IL-6-dependent mechanism that was augmented by, but not dependent on, IL-23 and was dependent on both transcription factors T-bet and AhR. Transfer of CD4(+) T cells differentiated with IL-6 in the absence of TGF-β ("Th22" cells) conferred complete protection of infected IL-22-deficient mice whereas transferred Th17 cells did not. These findings establish Th22 cells as an important component of mucosal antimicrobial host defense.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
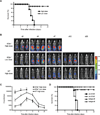
Figure 1
IL-23-independent, IL-22-dependent pathway to protection from C. rodentium Infection (A) Survival rates of _Il23a_−/− mice infected with high-dose (2 × 109 cfu) or low-dose (0.5 × 106 cfu) C. rodentium (n= 4–5 mice per group). Data are from one of three similar experiments. (B) Serial whole-body imaging of _Il23a_−/− mice inoculated with high- or low-dose of the luminescent strain of C. rodentium (strain ICC180) by gastric gavage and imaged at the indicated days PI. (C) Colonization kinetics of WT or _IL23a_−/− mice inoculated with indicated doses of C. rodentium. Light unit counts/sec directly correlate with the bacterial load (data not shown). Data are means +/− SEM. *p<0.01, **p<0.001. (D) Survival rates of _IL23a_−/− mice infected with low-dose C. rodentium and dosed IP with 150 µg of neutralizing anti-IL-22 mAb or isotype control every other day starting at d0 or d5. Data are representative of two or more experiments with a minimum of 3–4 mice per group.
Figure 2
IL-6 is indispensable for development of IL-22-producing CD4+ T cells (A) Naïve OTII-Tg CD4 T cells were sorted and activated by Ova peptide for 3d using splenic APCs from _Il23a_−/− or _Il6_−/− mice, with or without addition of the indicated neutralizing mAbs. Recovered cells were stained for surface CD4 and intracellular IL-22 and analyzed by flow cytometry. Numbers indicate the frequencies of IL-22+ CD4+ T cells. (B, C) Pooled frequency data from three or more independent experiments performed as in (A). Data are means +/− SEM (*p<0.05, **p<0.01).
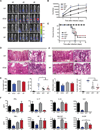
Figure 3
IL-23 governs early protective immunity to C. rodentium (A) Whole-body imaging of WT, _Il23a_−/−, _Il22_−/− and _Il6_−/− mice inoculated with 2 × 109 cfu of luminescent strain of C. rodentium and imaged at the indicated days PI. (B) Colonization kinetics of WT, _Il23a_−/−, _Il22_−/− and _Il6_−/− mice inoculated with 2 × 109 cfu of luminescent strain of C. rodentium. Data are means +/− SEM. *p<0.05, **p<0.01. (C) Survival rates of WT, _Il23a_−/−, _Il22_−/− and _Il6_−/− mice inoculated with 2 × 109 cfu of C. rodentium. (D) Histopathology of distal colonic tissues from WT, _Il23a_−/−, _Il22_−/− and _Il6_−/− mice collected eight days PI. H&E-stained sections (scale bars: 100 mm, left panel; 25 mm, right panels). White arrows denote individual clusters of C. rodentium attached to the apical surface of the colonic epithelium (WT) or dense colonies that extend into colonic crypts and penetrate the epithelium (_Il22_−/− and _Il6_−/−) or deeply invade the necrotic mucosa (_Il23a_−/−). Black arrows denote dying colonic epithelial cells. (E) Histopathological scoring of colons from infected WT, _Il23a_−/−, _Il22_−/− and _Il6_−/− mice was performed at d8 PI as per Supplemental Experimental Procedures. Data are means +/− SEM from two or more experiments (n=3–4 mice per group). *p<0.05, **p<0.01. (F) Multiplexed cytokine ELISA of the indicated cytokines and chemokines from supernatants of colonic tissue homogenates from WT, _Il23a_−/−, _Il6_−/−, _Il22_−/− mice collected d3 PI (24h cultures). Data are means +/− SEM from two or more experiments (n=3–4 mice per group). *p<0.05, **p<0.01.
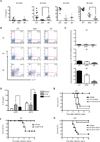
Figure 4
IL-6–induced CD4+ T cells are major source of protective IL-22 late in C. rodentium infection (A) WT, _Il23a_−/− and _Il6_−/− mice were inoculated with 2 × 109 cfu C. rodentium and colonic tissues collected at the indicated days PI for assessment of production of IL-22 in homogenate supernatants cultured for 24h. Data are means +/− SEM (*p<0.05, **p<0.01). (B) Colonic LP cells of WT, _Il23a_−/− and _Il6_−/− mice infected with C. rodentium were isolated on the indicated days PI and analyzed by flow cytometry for expression of intracellular IL-22 and IL-17A by CD3+CD4+ gated cells. Numbers indicate the frequencies of cells in each quadrant. (C) Flow cytometric frequency data for LPL isolates of WT, _Il23a_−/− and _Il6_−/− mice generated as in (B) and pooled from three independent experiments (n=3–4 mice per group). Data are means +/− SEM (*p<0.05, **p<0.01). (D) Survival rates of C. rodentium infected WT mice that were untreated or treated with neutralizing IL-22 mAb on days 0 and 3 PI, or on days 6, 7 and 8 PI. (E) Survival rates of _Il6_−/− mice infected with C. rodentium and treated with rIL-22-Fc protein (50 µg IP) or vehicle alone at d5 PI. (F) Survival rates of C. rodentium infected _Il22_−/− mice treated with rIL-22Fc protein (50 µg IP) or vehicle alone at days 0 and 3, or days 5 and 7 PI. (G) Survival rates of C. rodentium infected _Il22_−/− mice treated with rIL-22Fc protein (50 µg IP) or vehicle alone at days 0 and 3, or days 5 and 7 PI. All data are representative of three or more experiments with a minimum of 3–4 mice per group.
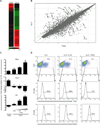
Figure 5
Th22 and Th17 cells are distinguished by reciprocal expression of T-bet, AhR and RORγt (A, B) FACS-sorted naïve OTII-Tg CD4+ T cells were activated by Ova peptide in presence of CD4+ T-cell depleted splenic APCs from _Il23a_−/− mice in the presence of IL-6 alone (Th22 polarization) or IL-6+TGFβ Thi7 polarization. CD4+ T cells were purified by magnetic sorting after 60h of culture and RNA was isolated for microarray-based gene expression analysis. Shown is a heat map of genes with more than 4-fold expression differences between the two populations (A) and transcriptome profiling of Th22- or Th17-polarized cells by scatterplot analysis (B). (C) Naïve CD4+ T cells isolated from C57BL/6 mice were activated by anti-CD3 and anti-CD28 in presence of IL-6 and IL-23 (Th22 polarization), IL-6 and the indicated concentrations of TGFβ Thi7 polarization), or absence of added cytokines (Th0). On day 3 the cells were further stimulated with PMA/ionomycin for 6 hours and analyzed by RT-PCR for Rorc ,Tbx21 and Ahr transcripts. Data are means +/− SEM (*p<0.05, **p<0.01). (D) Naïve OTII CD4+ T cells were simulated with Ova peptide in presence of IL-6, IL-6+IL-23 or IL-6+TGFβ and analyzed by flow cytometry for frequencies of IL-22+ CD4+ T cells on day 3 following stimulation for 4 hours with PMA/I in the presence of brefeldin A. The expression of T-bet and RORγt within the IL-22+ CD4+ gates was determined by intracellular staining (histograms). All data are representative of three or more experiments with a minimum of 3 mice per group.
Figure 6
IL-22 production by Th22 cells is T-bet- and AhR-dependent (A-C) FACS-sorted naïve CD4 T cells (CD4+CD25-CD62Lhigh CD44low) from WT, _Tbx21_−/− or _Ahr_−/− mice were activated with anti-CD3/CD28 in presence of IL-6 and IL-23 (Th22 polarization), IL-6 and different concentrations of TGFβ Thi7 polarizations, or absence of cytokines (Th0). On day 3 CD4+ T cells were re-stimulated with PMA/ionomycin for 6 hours and analyzed by RT-PCR for Il22 (A), Ahr and Tbx21 (B), or maf (C) transcripts. Expression values are normalized to Th0 controls. Data are means +/− SEM (*p<0.05). (D) Naïve CD4 T cells from WT or _Tbx21_−/− mice were differentiated under Th0, Th22 or Th17 conditions. Where indicated, the AhR-antagonist, CH-223191 (3µM), was added at the initiation of cultures. IL-22 protein was quantified by ELISA from culture supernatants of T cells treated with PMA/ionomycin for 48 hours. Data are means +/− SEM (*p<0.01). (E, F) WT and _Tbx21_−/− mice were inoculated with C. rodentium and sacrificed on d8 PI for analysis of IL-22 production from supernatants of colon homogenates cultured ex vivo for 24h (E; *p<0.05), or for analysis of expression of intracellular IL-22 and IFN-γ by CD3+CD4+ cells (F). Numbers in each quadrant indicate frequencies (CD3 and CD4 gated). (G) Colonization kinetics of WT _and Tbx21_−/− mice infected with 2 × 109 luminescent C. rodentium bacterial strain and assessed by whole body imaging on the indicated days PI. Data are means +/− SEM (*p<0.05, **p<0.01). All data are representative of two or more experiments.
Figure 7
Th22 cells provide superior host protection to Th17 cells (A, B) Naïve CD4+ T cells from WT mice were stimulated in vitro for 72h with anti-CD3+anti-CD28 with addition of IL-6 (Th22), IL-23 (IL-23 alone), or IL-6+TGFβ, without or with IL-23 addition (Th17 and Th17+IL-23, respectively), prior to isolation and transfer (A). Naïve CD4+ T cells from WT, _Il22_−/−, _Tbx21_−/− and _Ahr_−/− mice were similarly stimulated in vitro with addition of IL-6 (WT Th22, _Il22_−/− Th22, _Tbx21_−/− Th22 and _Ahr_−/− Th22, respectively), prior to isolation and transfer (B). N ≥ 5 mice per group; representative of two or more experiments.
Comment in
- IL-22 from T cells: better late than never.
Honda K. Honda K. Immunity. 2012 Dec 14;37(6):952-4. doi: 10.1016/j.immuni.2012.11.006. Immunity. 2012. PMID: 23244715
Similar articles
- STAT3 activation in Th17 and Th22 cells controls IL-22-mediated epithelial host defense during infectious colitis.
Backert I, Koralov SB, Wirtz S, Kitowski V, Billmeier U, Martini E, Hofmann K, Hildner K, Wittkopf N, Brecht K, Waldner M, Rajewsky K, Neurath MF, Becker C, Neufert C. Backert I, et al. J Immunol. 2014 Oct 1;193(7):3779-91. doi: 10.4049/jimmunol.1303076. Epub 2014 Sep 3. J Immunol. 2014. PMID: 25187663 - The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells.
Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. Qiu J, et al. Immunity. 2012 Jan 27;36(1):92-104. doi: 10.1016/j.immuni.2011.11.011. Epub 2011 Dec 15. Immunity. 2012. PMID: 22177117 Free PMC article. - Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection.
Guo X, Qiu J, Tu T, Yang X, Deng L, Anders RA, Zhou L, Fu YX. Guo X, et al. Immunity. 2014 Jan 16;40(1):25-39. doi: 10.1016/j.immuni.2013.10.021. Epub 2014 Jan 9. Immunity. 2014. PMID: 24412612 Free PMC article. - IL-17 and Th17 Cells.
Korn T, Bettelli E, Oukka M, Kuchroo VK. Korn T, et al. Annu Rev Immunol. 2009;27:485-517. doi: 10.1146/annurev.immunol.021908.132710. Annu Rev Immunol. 2009. PMID: 19132915 Review. - Th17 cells and mucosal host defense.
Aujla SJ, Dubin PJ, Kolls JK. Aujla SJ, et al. Semin Immunol. 2007 Dec;19(6):377-82. doi: 10.1016/j.smim.2007.10.009. Epub 2007 Nov 28. Semin Immunol. 2007. PMID: 18054248 Free PMC article. Review.
Cited by
- The Th17 family: flexibility follows function.
Basu R, Hatton RD, Weaver CT. Basu R, et al. Immunol Rev. 2013 Mar;252(1):89-103. doi: 10.1111/imr.12035. Immunol Rev. 2013. PMID: 23405897 Free PMC article. Review. - Long-term use of broad-spectrum antibiotics affects Ly6Chi monocyte recruitment and IL-17A and IL-22 production through the gut microbiota in tumor-bearing mice treated with chemotherapy.
Wu Y, Tang X, Hu F, Zhu T, Liu H, Xiong Y, Zuo X, Xu A, Zhuang X. Wu Y, et al. Immunol Res. 2022 Dec;70(6):829-843. doi: 10.1007/s12026-022-09313-9. Epub 2022 Sep 23. Immunol Res. 2022. PMID: 36149530 - Activation of the aryl hydrocarbon receptor in inflammatory bowel disease: insights from gut microbiota.
Hou JJ, Ma AH, Qin YH. Hou JJ, et al. Front Cell Infect Microbiol. 2023 Oct 24;13:1279172. doi: 10.3389/fcimb.2023.1279172. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37942478 Free PMC article. Review. - Functions of Murine Dendritic Cells.
Durai V, Murphy KM. Durai V, et al. Immunity. 2016 Oct 18;45(4):719-736. doi: 10.1016/j.immuni.2016.10.010. Immunity. 2016. PMID: 27760337 Free PMC article. Review. - IL-23-mediated mononuclear phagocyte crosstalk protects mice from Citrobacter rodentium-induced colon immunopathology.
Aychek T, Mildner A, Yona S, Kim KW, Lampl N, Reich-Zeliger S, Boon L, Yogev N, Waisman A, Cua DJ, Jung S. Aychek T, et al. Nat Commun. 2015 Mar 12;6:6525. doi: 10.1038/ncomms7525. Nat Commun. 2015. PMID: 25761673 Free PMC article.
References
- Andoh A, Zhang Z, Inatomi O, Fujino S, Deguchi Y, Araki Y, Tsujikawa T, Kitoh K, Kim-Mitsuyama S, Takayanagi A, et al. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969–984. - PubMed
- Bry L, Brenner MB. Critical role of T cell-dependent serum antibody, but not the gut-associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. J. Immunol. 2004;172:433–441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 DK071176/DK/NIDDK NIH HHS/United States
- P30 AR048311/AR/NIAMS NIH HHS/United States
- R01 DK093015/DK/NIDDK NIH HHS/United States
- R24 DK064400/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials