Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing - PubMed (original) (raw)
Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing
Nicholas A Bokulich et al. Nat Methods. 2013 Jan.
Abstract
High-throughput sequencing has revolutionized microbial ecology, but read quality remains a considerable barrier to accurate taxonomy assignment and α-diversity assessment for microbial communities. We demonstrate that high-quality read length and abundance are the primary factors differentiating correct from erroneous reads produced by Illumina GAIIx, HiSeq and MiSeq instruments. We present guidelines for user-defined quality-filtering strategies, enabling efficient extraction of high-quality data and facilitating interpretation of Illumina sequencing results.
Figures
Figure 1
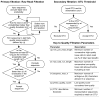
Quality Filtration Process Flow in QIIME v1.5.0.
Figure 2
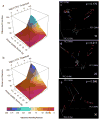
α- and β-Diversity comparisons of mock community reads filtered using select phred_quality_score (q) settings (dataset 1). A, B: Family-level (A) and genus-level (B) taxon counts for mock communities filtered with variable (q) values at multiple OTU minimum abundance thresholds (c) (as %). Arrows below color key indicate expected genus- (blue) and family-level (red) taxon counts. C, D, E: Procrustes PCoA biplot of GAIIx weighted UniFrac distance comparing variation in (q). Comparison of (q) setting listed in bottom-right corner to (q) = 3. Top-right corner indicates Bonferroni-corrected _p_-value for Procrustes goodness of fit. Red, human feces; Magenta, mock community; Cyan, human skin; Dark cyan, human tongue; Blue, freshwater; Orange, freshwater creek; Purple, ocean; Yellow, estuary sediment; Pink, soil. All other settings represent defaults in both α- and β-diversity comparisons.
References
- Gilbert JA, Meyer F. ASM Microbe Magazine. Mar, 2012. 2012.
- Quince C, et al. Nat Methods. 2009;6:639–644. - PubMed
- Caporaso JG, et al. PNAS. 2011;108:4516–4522. - PubMed
Publication types
MeSH terms
Grants and funding
- U54HG004969/HG/NHGRI NIH HHS/United States
- P01 DK078669/DK/NIDDK NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01HD059127/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U54 HG004969/HG/NHGRI NIH HHS/United States
- R01 HD059127/HD/NICHD NIH HHS/United States
- DK78669/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases