STRING v9.1: protein-protein interaction networks, with increased coverage and integration - PubMed (original) (raw)
. 2013 Jan;41(Database issue):D808-15.
doi: 10.1093/nar/gks1094. Epub 2012 Nov 29.
Affiliations
- PMID: 23203871
- PMCID: PMC3531103
- DOI: 10.1093/nar/gks1094
STRING v9.1: protein-protein interaction networks, with increased coverage and integration
Andrea Franceschini et al. Nucleic Acids Res. 2013 Jan.
Abstract
Complete knowledge of all direct and indirect interactions between proteins in a given cell would represent an important milestone towards a comprehensive description of cellular mechanisms and functions. Although this goal is still elusive, considerable progress has been made-particularly for certain model organisms and functional systems. Currently, protein interactions and associations are annotated at various levels of detail in online resources, ranging from raw data repositories to highly formalized pathway databases. For many applications, a global view of all the available interaction data is desirable, including lower-quality data and/or computational predictions. The STRING database (http://string-db.org/) aims to provide such a global perspective for as many organisms as feasible. Known and predicted associations are scored and integrated, resulting in comprehensive protein networks covering >1100 organisms. Here, we describe the update to version 9.1 of STRING, introducing several improvements: (i) we extend the automated mining of scientific texts for interaction information, to now also include full-text articles; (ii) we entirely re-designed the algorithm for transferring interactions from one model organism to the other; and (iii) we provide users with statistical information on any functional enrichment observed in their networks.
Figures
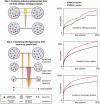
Figure 1.
Improved procedure for interaction transfer between organisms. Left: steps 1 and 2 of the functional association transfer pipeline. In the first step, the individual links between proteins are combined into a score between orthologous groups, sequentially, from the strongest link (thick line) to the weakest (thin). Each subsequent score is down-weighted, both based on the similarity of its organism to organisms that have already contributed to the combined scores, and on number of proteins from the same organism inside the orthologous group. In the second step of the transfer pipeline, the links between orthologous groups are transferred back to individual protein pairs belonging to these groups. This is done sequentially from the lowest to highest taxonomy level. In the above example, the two transferred links from the highest taxonomic level (orange links) are penalized for the increase in number of proteins from the target species in one of the orthologous groups. Right: ROC curves indicating the performance of predicted interolog scores, benchmarked against KEGG pathways; an inferred link between two proteins is considered to be a true positive when both proteins are annotated to be together in at least one shared KEGG pathway.
Figure 2.
Network visualization and statistical analysis of a user-supplied protein list. The STRING screenshot shows a user-supplied set of genes, here a selection of cancer genes as annotated at the COSMIC database (52). The set is restricted to those genes that are known to pre-dispose to cancer already when mutated in the germline, and that have at least one connection in STRING. The inset illustrates the website’s new functionality for automatically detecting statistically enriched functions or processes in a network. In this example, one of the detected processes (nucleotide excision repair) is of interest and has been selected; STRING automatically highlighted the corresponding nodes in the network, where they are seen to form a densely connected module.
Similar articles
- The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets.
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. Szklarczyk D, et al. Nucleic Acids Res. 2021 Jan 8;49(D1):D605-D612. doi: 10.1093/nar/gkaa1074. Nucleic Acids Res. 2021. PMID: 33237311 Free PMC article. - The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored.
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. Szklarczyk D, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D561-8. doi: 10.1093/nar/gkq973. Epub 2010 Nov 2. Nucleic Acids Res. 2011. PMID: 21045058 Free PMC article. - STRING v10: protein-protein interaction networks, integrated over the tree of life.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. Szklarczyk D, et al. Nucleic Acids Res. 2015 Jan;43(Database issue):D447-52. doi: 10.1093/nar/gku1003. Epub 2014 Oct 28. Nucleic Acids Res. 2015. PMID: 25352553 Free PMC article. - Merging in-silico and in vitro salivary protein complex partners using the STRING database: A tutorial.
Crosara KTB, Moffa EB, Xiao Y, Siqueira WL. Crosara KTB, et al. J Proteomics. 2018 Jan 16;171:87-94. doi: 10.1016/j.jprot.2017.08.002. Epub 2017 Aug 3. J Proteomics. 2018. PMID: 28782718 Review. - Databases of protein-protein interactions and complexes.
Ooi HS, Schneider G, Chan YL, Lim TT, Eisenhaber B, Eisenhaber F. Ooi HS, et al. Methods Mol Biol. 2010;609:145-59. doi: 10.1007/978-1-60327-241-4_9. Methods Mol Biol. 2010. PMID: 20221918 Review.
Cited by
- Deciphering molecular landscape of breast cancer progression and insights from functional genomics and therapeutic explorations followed by in vitro validation.
Khan B, Qahwaji R, Alfaifi MS, Athar T, Khan A, Mobashir M, Ashankyty I, Imtiyaz K, Alahmadi A, Rizvi MMA. Khan B, et al. Sci Rep. 2024 Nov 20;14(1):28794. doi: 10.1038/s41598-024-80455-6. Sci Rep. 2024. PMID: 39567714 Free PMC article. - VI-VS: calibrated identification of feature dependencies in single-cell multiomics.
Boyeau P, Bates S, Ergen C, Jordan MI, Yosef N. Boyeau P, et al. Genome Biol. 2024 Nov 15;25(1):294. doi: 10.1186/s13059-024-03419-z. Genome Biol. 2024. PMID: 39548591 Free PMC article. - Dataset of Panda sperm proteome.
Liu S, Wang T, Liu Y, Wang S, Li F, Chen J, Hu X, Zhang M, Wang J, Li Y, James A, Hou R, Cai K. Liu S, et al. Data Brief. 2024 Oct 21;57:111052. doi: 10.1016/j.dib.2024.111052. eCollection 2024 Dec. Data Brief. 2024. PMID: 39525650 Free PMC article. - Striatal dopamine gene network moderates the effect of early adversity on the risk for adult psychiatric and cardiometabolic comorbidity.
Barth B, Arcego DM, de Mendonça Filho EJ, de Lima RMS, Parent C, Dalmaz C, Portella AK, Pokhvisneva I, Meaney MJ, Silveira PP. Barth B, et al. Sci Rep. 2024 Nov 9;14(1):27349. doi: 10.1038/s41598-024-78465-5. Sci Rep. 2024. PMID: 39521843 Free PMC article. - Identifying the HIV-Resistance-Related Factors and Regulatory Network via Multi-Omics Analyses.
Long X, Liu G, Liu X, Zhang C, Shi L, Zhu Z. Long X, et al. Int J Mol Sci. 2024 Nov 1;25(21):11757. doi: 10.3390/ijms252111757. Int J Mol Sci. 2024. PMID: 39519306 Free PMC article.
References
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–544. - PubMed
- Wolf YI, Grishin NV, Koonin EV. Estimating the number of protein folds and families from complete genome data. J.Mol. Biol. 2000;299:897–905. - PubMed
- Aloy P, Russell RB. Ten thousand interactions for the molecular biologist. Nature Biotechnol. 2004;22:1317–1321. - PubMed
- Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405:823–826. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous