Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae - PubMed (original) (raw)
. 2013 Jan;41(2):1191-210.
doi: 10.1093/nar/gks1056. Epub 2012 Dec 2.
Juliane Merl, Martina Sauert, Jorge Perez-Fernandez, Patrick Schultz, Astrid Bruckmann, Stephan Hamperl, Uli Ohmayer, Reinhard Rachel, Anja Jacob, Kristin Hergert, Rainer Deutzmann, Joachim Griesenbeck, Ed Hurt, Philipp Milkereit, Jochen Baßler, Herbert Tschochner
Affiliations
- PMID: 23209026
- PMCID: PMC3553968
- DOI: 10.1093/nar/gks1056
Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae
Thomas Hierlmeier et al. Nucleic Acids Res. 2013 Jan.
Abstract
Eukaryotic ribosome biogenesis requires more than 150 auxiliary proteins, which transiently interact with pre-ribosomal particles. Previous studies suggest that several of these biogenesis factors function together as modules. Using a heterologous expression system, we show that the large ribosomal subunit (LSU) biogenesis factor Noc1p of Saccharomyces cerevisiae can simultaneously interact with the LSU biogenesis factor Noc2p and Rrp5p, a factor required for biogenesis of the large and the small ribosomal subunit. Proteome analysis of RNA polymerase-I-associated chromatin and chromatin immunopurification experiments indicated that all members of this protein module and a specific set of LSU biogenesis factors are co-transcriptionally recruited to nascent ribosomal RNA (rRNA) precursors in yeast cells. Further ex vivo analyses showed that all module members predominantly interact with early pre-LSU particles after the initial pre-rRNA processing events have occurred. In yeast strains depleted of Noc1p, Noc2p or Rrp5p, levels of the major LSU pre-rRNAs decreased and the respective other module members were associated with accumulating aberrant rRNA fragments. Therefore, we conclude that the module exhibits several binding interfaces with pre-ribosomes. Taken together, our results suggest a co- and post-transcriptional role of the yeast Rrp5p-Noc1p-Noc2p module in the structural organization of early LSU precursors protecting them from non-productive RNase activity.
Figures
Figure 1.
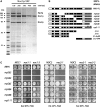
The ribosome biogenesis factors Noc1p, Noc2p and Rrp5p interact in vivo. (A) Noc1p-TAP efficiently co-purifies Noc2p and Rrp5p from yeast extracts. Noc1-TAP was isolated from extracts of strain Y3572 applying the tandem affinity purification method under increasing concentration of NaCl. The composition of the Noc1p containing particles was analysed on a 4–12% gradient SDS–PAGE gel, stained with Coomassie Blue. Prominent co-purified proteins are indicated. (B) Overview of the rrp5 alleles analysed in this study and schematic presentation of the corresponding protein variants. Black bars illustrate the S1 RNA binding motifs, grey bars the tetratricopeptide repeats, respectively [adapted from (20); see ‘Introduction’ section]. Point mutations in the temperature sensitive rrp5-11 allele are indicated by asterisk. (C) Genetic interaction studies show synthetic lethal effects between rrp5 alleles with deletions of N-terminal S1-repeats and _noc1_-ts or _noc2_-ts mutants. The respective double shuffle strains (Y3668, Y4207 and Y3715) were transformed with plasmids carrying the indicated wild-type and mutant alleles of ProtA-RRP5, ProtA-NOC1, NOC2 or ProtA-NOC3 (
Supplementary Figure S4
). Transformants were spotted in a 10-fold dilution series on SDC + FOA plates and incubated at the indicated temperature and for the indicated time to analyse genetic interactions.
Figure 2.
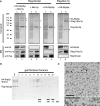
Reconstitution of the Rrp5p–Noc1p–Noc2p module from proteins co-expressed in insect cells. The indicated combinations of Flag-tagged bait proteins and potential interaction partners were co-expressed in SF21 insect cells infected with recombinant baculo viruses containing the plasmids TK1232, TK1504, TK1658 or TK1677 (
Supplementary Figure S4
). (A) The bait proteins were purified with anti-Flag affinity matrix from lysates of 50 × 106 infected cells and eluted with buffer containing Flag peptide. Aliquots of the lysates (L) and eluates (E) were analysed by coomassie stained SDS–PAGE (upper panel; 0.05% L, 23% E) and MS (HA-Rrp5p is indicated by “*”; Flag-Noc2p by “°”; (Flag-) Noc1p by “+”; N-terminal Flag-Noc1p fragment by “×”) or Western blotting using anti-HA, anti-Noc1p and anti-Flag antibodies (lower panel; 0.006% L, 1.2% E). (B) Affinity purified Flag-Noc2p–Noc1p–Rrp5p complex was fractionated on a Superose 6 gel filtration column. Aliquots of the eluate (E; 20%) and the fractions (2–20; 40%) were analysed by Coomassie stained SDS–PAGE. Elution of marker proteins in independent gel filtration runs is indicated at the bottom. (C) Electron micrograph of an uranyl acetate stained aliquot of the Flag-Noc2p–Noc1p–Rrp5p complex after Superose 6 gel filtration column from an independent purification (corresponding to fraction 10 in Figure 2B). Arrows and arrowheads indicate particles of 12 nm and 8 nm diameter, respectively. The lower panel shows an enlarged view of the boxed area. Scale bars are 70 nm.
Figure 3.
The N-terminal part of Rrp5p is required for stable association with Noc1p. (A) Co-precipitation of full-length and truncated Rrp5p variants with Noc1p from yeast cells. Genomic NOC1-TAP, rrp5Δ yeast strains harbouring plasmids encoding Rrp5p-FL-GFP (FL) or Rrp5p-S1-9-GFP and Rrp5p-S10-TPR-GFP (N + C; Figure 1B and
Supplementary Figure S4
) were generated by plasmid shuffling from strain Y3716 and used for tandem affinity purification of Noc1p. The calmodulin eluates were analysed by coomassie stained SDS–PAGE (left panel), whereas the supernatant of the cell lysate and calmodulin eluates were analysed by western blotting (right panel) using anti-GFP (upper panel) and anti-CBP antibodies (lower panel) to detect Rrp5-GFP variants or Noc1-CBP, respectively. (B) Heterologous reconstitution of complexes containing Flag-Noc1p and HA-Rrp5p variants. The indicated combinations of Flag-Noc1p and truncated HA-Rrp5p variants (Figure 1B) were co-expressed in SF21 insect cells infected with recombinant baculo viruses containing the plasmids TK1658, TK1727, TK1728, TK1729 or TK1732 (
Supplementary Figure S4
). Flag-Noc1p was purified with anti-Flag affinity matrix from lysates of 50 × 106 infected cells and eluted with buffer containing Flag peptide. Aliquots of the lysates (L) and eluates (E) were analysed by SDS–PAGE and coomassie staining (upper panel; 0.04% L, 20% E) or western blotting using anti-HA and anti-Flag antibodies (lower panels; 0.006% L, 1% E). Coomassie-stained proteins were identified by mass spectrometry (HA-Rrp5p variants are indicated by “*”; Flag-Noc1p by “+”; N-terminal Flag-Noc1p fragments by “×”).
Figure 4.
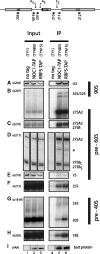
Comparison of pre-ribosomal particles associated with Noc1p or Rrp5p. Yeast strains expressing chromosomally encoded Noc1p-TAP (TY483) or Rrp5p-TAP (TY615) and an untagged control strain (TY1) were grown to exponential phase (OD600 = 0.8) in rich medium. TAP-tagged proteins were affinity purified from cell extracts using IgG-coupled magnetic beads. After washing, the beads were split for the analysis of co-purified RNAs and proteins. RNA isolated from aliquots of cell extracts (Input) and precipitates (IP) was separated on acryl amide (A, E) or agarose (B, C, F, G, H) gels and analysed by Northern blotting using the indicated probes (upper panel, o202–o1819). Alternatively, the isolated RNA was used as template in primer extension reactions (D). Purification of the bait proteins was controlled by Western blotting (I) against the Protein A moiety of the TAP tag using PAP detection reagent. Equal signal intensities in Input and IP correspond to 2 and 11% precipitation efficiency in Northern (0.067% In, 3.33% IP) and Western blot (0.11% In, 1% IP) analysis, respectively. (signal potentially arising from 27SA3 pre-rRNA is indicated by asterisk).
Figure 5.
Comparative proteome analysis of Noc1p and Rrp5p containing pre-ribosomes. Proteins from aliquots of Noc1p-TAP and Rrp5p-TAP purifications (Figure 4) were digested using trypsin and the resulting peptides were labelled with iTRAQ(R) reagents 116 (Noc1p purification) and 117 (Rrp5p purification), subsequently pooled, separated by reversed phase nano HPLC and spotted on a MALDI–MS/MS target. The top eight peptides of the MS run in each spot were selected for fragmentation in MS/MS mode to determine the sequence of the peptide and the ratio of the iTRAQ reporter group signal intensities (117/116). For proteins identified with more than one peptide [ion score confidence interval (CI) >95%] the average iTRAQ ratio (117/116 = RRP5/NOC1) was calculated. LSU biogenesis factors (A) and SSU processome components (B) identified in both purifications are listed according to their average iTRAQ ratio (error bars are standard deviations of the average ratio; numbers of identified peptides are indicated in brackets). The average iTRAQ ratios of ribosomal proteins of the large (rpL) and the small (rpS) subunit identified with more than one peptide are shown in (A). (C) Peptide count analysis of RNA polymerase subunits identified in both purifications. Only peptides of Pol-I-specific subunits (A12.2, A49, A135, A190) and of common subunits (AC40, ABC23) were identified with a score >95% CI (one peptide each).
Figure 6.
A specific set of LSU and SSU biogenesis factors is part of Pol-I-transcribed chromatin. Yeast strains expressing chromosomally encoded Protein A fusion proteins of the Pol-I subunit Rpa135p (TY2423) or the Pol-II subunit Rpb2p (TY2424) were grown in rich medium to exponential phase (OD600 ∼0.5–0.8) and cross linked using formaldehyde. Chromatin fractions were prepared and the bait proteins were purified using IgG coupled magnetic beads. The co-purified proteins were subjected to comparative MS analysis using iTRAQ reagents. For proteins identified with at least one peptide with an ion score of >95% CI, the (average) iTRAQ ratio (Pol-II versus Pol-I purification) was calculated to determine the relative abundance of the respective protein in the Pol-I purification compared with the Pol-II purification. The results of six independent experiments were subjected to statistical analysis using clustering algorithms to determine groups of proteins specifically associated with Pol-I or Pol-II-transcribed chromatin. Only proteins identified in at least four out of six experiments were included. (A) Overview of the cluster analysis of six independent comparative Pol-II/Pol-I purifications with the five main clusters indicated on the right. The colour code for the log2 transformed iTRAQ ratios of Pol-II versus Pol-I purifications are indicated (for details, see ‘Materials and Methods’ section). grey: protein not identified in this experiment; blue: enriched in Pol-I purifications; yellow: enriched in Pol-II purifications; black: equally abundant in Pol-I and Pol-II purifications. (B, C, E, F) Detailed view of the clusters A, B, D and E, respectively, including a classification of the identified proteins. rDNA: shown to co-immunoprecipitate rDNA; Pol1: Pol-I subunit; SSU/Sno: part of SSU-processome/90S pre-ribosome or snoRNPs involved in ribosome biogensis; LSU: Large ribosomal subunit biogenesis factor; mRNP: part of co-transcriptionally formed mRNP; elongation: Pol-II elongation factor; Pol2-assoc.: shown to associate with Pol-II. LSU biogenesis factors are shown in red if they were already identified in the Noc1p/Rrp5p proteome analyses shown in Figure 4. Identified components of the Rrp5p–Noc1p–Noc2p module are indicated by asterisk. (D) Proteins in cluster C are classified and listed according to their physiological function. The numbers of identified proteins are indicated in brackets.
Figure 7.
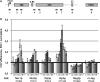
Analysis of co-transcriptional recruitment of Noc1p, Noc2p and Rrp5p to the 35S rRNA gene. (A) Primer pair positions on the rRNA gene to perform quantitative PCR (qPCR) analysis of ChIP experiments. The relative positions of the rDNA amplicons analysed by qPCR (1–8) are indicated. For normalization, an amplicon in the PDC1 gene (9) was used. (B) ChIP experiments were performed with yeast strains in which Noc1p (TY483), Noc2p (TY577), Rrp5p (TY615), Utp4p (TY1540) or Nog2p (TY1965) are expressed as tandem affinity purification (TAP) tag fusion proteins, and with a control strain expressing no tagged protein (TY543). Cells were grown in rich medium at 30°C to exponential phase (OD600 = 0.5–0.7) and cross-linked using formaldehyde (final concentration 1%) for 15’ at 30°C. ChIP analysis was performed as described in ‘Materials and Methods’ section. The amounts of specific DNA fragments present in the input and retained on the beads were determined by qPCR with primer pairs amplifying the regions 1–8 of the rDNA depicted in the schematic representation and of the PDC1 gene (primer pair 9). In each experiment the precipitation efficiencies [percent IP (rDNA)] for the respective amplified DNA regions were calculated and normalized to the PDC1 precipitation efficiencies [percent IP (rDNA)/percent IP (PDC1)]. The graph shows the average of three biological replicates, including standard deviations. A black line depicts the internal background as a result of the normalization to the precipitated PDC1 DNA.
Figure 8.
Analysis of the binding hierarchy of biogenesis factors to pre-ribosomal particles. Yeast strains expressing chromosomally encoded Noc1p-TAP (TY2302/TY2343) or Noc2p-TAP (TY2303/TY2301) in which the RRP5 gene is either under control of the endogenous or the inducible GAL 1/10 promoter were cultivated for 10 and 18 h in glucose containing rich medium (final OD600 = 0.5–1). Analogous experiments were carried out with strains expressing chromosomally encoded Rrp5p-TAP (TY2499/TY2501) or Utp22p-TAP (TY2417/TY2418) with NOC1 under the control of its endogenous or the GAL 1/10 promoter, and for strains that express chromosomally encoded Rrp5p-TAP (TY2500/TY2502) with NOC2 under the control of its endogenous or the GAL1/10 promoter. These strains were cultivated for 10,18 and 24 h in glucose containing rich medium (final OD600 = 0.5–1). The respective background strains expressing no tagged protein (TY1, TY616), which served as controls, were cultivated for 18 h in glucose containing rich medium. TAP-tagged proteins were affinity purified from cell extracts using IgG sepharose. After washing, the beads were split for the analysis of co-purified RNAs and proteins. RNAs isolated from aliquots of cell extracts (Input) and immuno-purified fractions (IP) were separated on acrylamide or agarose gels and analysed by Northern blotting by subsequent hybridization using the indicated probes (o202–o1819; binding sites are depicted in
Supplementary Figure S1A
). In general, all input and IP samples of the purification of one bait protein in the different strains were analysed on the same gel [see also entire blots shown in the
Supplementary Material
(
Supplementary Figure S7
)]. Purification of the bait proteins was controlled by Western blotting (WB) against the Protein A moiety of the TAP tag using anti-Protein A antibody. (A) Depletion of Rrp5p; (B) depletion of Noc2p; (C) depletion of Noc1p; (D) control strains. Equal signal intensities in Input and IP correspond to 2 and 1.5% purification efficiency in Northern blots of agarose and acrylamide gels, respectively. In Western blots, equal signal intensities in Input and IP correspond to 20% (Rrp5-TAP), 33% (Noc1-TAP), 17% (Noc2-TAP) and 50% (UTP22-TAP) purification efficiencies. Aberrant pre-rRNA fragments resulting from depletion of biogenesis factors are indicated by multiplication symbol.
Similar articles
- Rrp5p, a trans-acting factor in yeast ribosome biogenesis, is an RNA-binding protein with a pronounced preference for U-rich sequences.
de Boer P, Vos HR, Faber AW, Vos JC, Raué HA. de Boer P, et al. RNA. 2006 Feb;12(2):263-71. doi: 10.1261/rna.2257606. RNA. 2006. PMID: 16428605 Free PMC article. - Studies on the Coordination of Ribosomal Protein Assembly Events Involved in Processing and Stabilization of Yeast Early Large Ribosomal Subunit Precursors.
Ohmayer U, Gil-Hernández Á, Sauert M, Martín-Marcos P, Tamame M, Tschochner H, Griesenbeck J, Milkereit P. Ohmayer U, et al. PLoS One. 2015 Dec 7;10(12):e0143768. doi: 10.1371/journal.pone.0143768. eCollection 2015. PLoS One. 2015. PMID: 26642313 Free PMC article. - The Npa1p complex chaperones the assembly of the earliest eukaryotic large ribosomal subunit precursor.
Joret C, Capeyrou R, Belhabich-Baumas K, Plisson-Chastang C, Ghandour R, Humbert O, Fribourg S, Leulliot N, Lebaron S, Henras AK, Henry Y. Joret C, et al. PLoS Genet. 2018 Aug 31;14(8):e1007597. doi: 10.1371/journal.pgen.1007597. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30169518 Free PMC article. - Joining the interface: a site for Nmd3 association on 60S ribosome subunits.
Oeffinger M. Oeffinger M. J Cell Biol. 2010 Jun 28;189(7):1071-3. doi: 10.1083/jcb.201006033. J Cell Biol. 2010. PMID: 20584913 Free PMC article. Review. - Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast.
Konikkat S, Woolford JL Jr. Konikkat S, et al. Biochem J. 2017 Jan 15;474(2):195-214. doi: 10.1042/BCJ20160516. Biochem J. 2017. PMID: 28062837 Free PMC article. Review.
Cited by
- Disruption of ribosome assembly in yeast blocks cotranscriptional pre-rRNA processing and affects the global hierarchy of ribosome biogenesis.
Talkish J, Biedka S, Jakovljevic J, Zhang J, Tang L, Strahler JR, Andrews PC, Maddock JR, Woolford JL Jr. Talkish J, et al. RNA. 2016 Jun;22(6):852-66. doi: 10.1261/rna.055780.115. Epub 2016 Apr 1. RNA. 2016. PMID: 27036125 Free PMC article. - Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae.
Ohmayer U, Gamalinda M, Sauert M, Ossowski J, Pöll G, Linnemann J, Hierlmeier T, Perez-Fernandez J, Kumcuoglu B, Leger-Silvestre I, Faubladier M, Griesenbeck J, Woolford J, Tschochner H, Milkereit P. Ohmayer U, et al. PLoS One. 2013 Jul 10;8(7):e68412. doi: 10.1371/journal.pone.0068412. Print 2013. PLoS One. 2013. PMID: 23874617 Free PMC article. - Pol5 is required for recycling of small subunit biogenesis factors and for formation of the peptide exit tunnel of the large ribosomal subunit.
Braun CM, Hackert P, Schmid CE, Bohnsack MT, Bohnsack KE, Perez-Fernandez J. Braun CM, et al. Nucleic Acids Res. 2020 Jan 10;48(1):405-420. doi: 10.1093/nar/gkz1079. Nucleic Acids Res. 2020. PMID: 31745560 Free PMC article. - NOC1 is a direct MYC target, and its protein interactome dissects its activity in controlling nucleolar function.
Manara V, Radoani M, Belli R, Peroni D, Destefanis F, Angheben L, Tome G, Tebaldi T, Bellosta P. Manara V, et al. Front Cell Dev Biol. 2023 Dec 28;11:1293420. doi: 10.3389/fcell.2023.1293420. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38213308 Free PMC article. - Discovery of Enzymatic Targets of Transcriptional Activators via in Vivo Covalent Chemical Capture.
Dugan A, Majmudar CY, Pricer R, Niessen S, Lancia JK, Fung HY, Cravatt BF, Mapp AK. Dugan A, et al. J Am Chem Soc. 2016 Sep 28;138(38):12629-35. doi: 10.1021/jacs.6b07680. Epub 2016 Sep 20. J Am Chem Soc. 2016. PMID: 27611834 Free PMC article.
References
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. - PubMed
- Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. - PubMed
- Mougey EB, O’Reilly M, Osheim Y, Miller OL, Jr, Beyer A, Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 1993;7:1609–1619. - PubMed
- Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases