Bioinformatics for the Human Microbiome Project - PubMed (original) (raw)
Bioinformatics for the Human Microbiome Project
Dirk Gevers et al. PLoS Comput Biol. 2012.
No abstract available
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
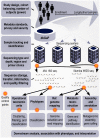
Figure 1. Bioinformatics in the HMP as a model for further studies of the human microbiome.
Important computational considerations throughout the design, implementation, and analysis of a large human microbiome study such as the HMP; for details of the HMP's specific computational protocols, see , . In the HMP, study design considerations included cohort balancing for gender and geographic location and recruitment of 300 individuals for adequate power. Subject metadata were protected and distributed through dbGaP , and up to three longitudinal samples were drawn from the microbiomes of 18 body habitats. These were tracked and sequenced at up to four distinct centers, including >5,000 16S rRNA gene datasets using 454 reads from the V1–3 and V3–5 hypervariable regions and >700 Illumina whole-genome shotgun datasets totaling over 8 Tbp of sequence. Quality control of sequences and datasets was performed at multiple points throughout data generation. Computational pipelines were developed and documented for each sequence data product as well as downstream analyses, with full results and protocols available at the HMP Data Analysis and Coordinating Center (
).
Figure 2. Topics in the study of the human microbiome with outstanding computational biology challenges.
There remain many areas in the study of the human microbiome that will benefit from further bioinformatic efforts. At a whole-population level, the dynamics and stochasticity of microbiome acquisition at birth and its subsequent intersubject transmission must be characterized. As individual hosts, we each expose our microbiomes to unique genetic, dietary, pharmaceutical, and environmental perturbations, which in turn dictate systematic immune responses that are governed by individual sensing and regulatory biomolecular mechanisms. Within our microbiome, both host-microbe and microbe-microbe interactions dictate community ecology. These are governed by a variety of molecular mechanisms well-studied in model microbes including protein–protein interactions, metabolism, regulatory networks, and extracellular transport. In many of the most difficult assay types, such as whole-community proteomics or metabolomics, informatic challenges such as molecular identification remain to be overcome.
Similar articles
- MetCap: a bioinformatics probe design pipeline for large-scale targeted metagenomics.
Kushwaha SK, Manoharan L, Meerupati T, Hedlund K, Ahrén D. Kushwaha SK, et al. BMC Bioinformatics. 2015 Feb 28;16(1):65. doi: 10.1186/s12859-015-0501-8. BMC Bioinformatics. 2015. PMID: 25880302 Free PMC article. - Use of whole genome shotgun metagenomics: a practical guide for the microbiome-minded physician scientist.
Ma J, Prince A, Aagaard KM. Ma J, et al. Semin Reprod Med. 2014 Jan;32(1):5-13. doi: 10.1055/s-0033-1361817. Epub 2014 Jan 3. Semin Reprod Med. 2014. PMID: 24390915 Review. - Databases of the marine metagenomics.
Mineta K, Gojobori T. Mineta K, et al. Gene. 2016 Feb 1;576(2 Pt 1):724-8. doi: 10.1016/j.gene.2015.10.035. Epub 2015 Oct 28. Gene. 2016. PMID: 26518717 Review. - Metagenomic pyrosequencing and microbial identification.
Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Petrosino JF, et al. Clin Chem. 2009 May;55(5):856-66. doi: 10.1373/clinchem.2008.107565. Epub 2009 Mar 5. Clin Chem. 2009. PMID: 19264858 Free PMC article. Review. - [Bioinformatics, systems biology and KEGG].
Honda W, Tanabe M, Yano A, Kanehisa M. Honda W, et al. Seikagaku. 2008 Dec;80(12):1094-111. Seikagaku. 2008. PMID: 19172790 Review. Japanese. No abstract available.
Cited by
- The vaginal mycobiome: A contemporary perspective on fungi in women's health and diseases.
Bradford LL, Ravel J. Bradford LL, et al. Virulence. 2017 Apr 3;8(3):342-351. doi: 10.1080/21505594.2016.1237332. Epub 2016 Sep 22. Virulence. 2017. PMID: 27657355 Free PMC article. Review. - IM-TORNADO: a tool for comparison of 16S reads from paired-end libraries.
Jeraldo P, Kalari K, Chen X, Bhavsar J, Mangalam A, White B, Nelson H, Kocher JP, Chia N. Jeraldo P, et al. PLoS One. 2014 Dec 15;9(12):e114804. doi: 10.1371/journal.pone.0114804. eCollection 2014. PLoS One. 2014. PMID: 25506826 Free PMC article. - How we decide what to eat: Toward an interdisciplinary model of gut-brain interactions.
Plassmann H, Schelski DS, Simon MC, Koban L. Plassmann H, et al. Wiley Interdiscip Rev Cogn Sci. 2022 Jan;13(1):e1562. doi: 10.1002/wcs.1562. Epub 2021 May 11. Wiley Interdiscip Rev Cogn Sci. 2022. PMID: 33977675 Free PMC article. Review. - A Review on the Protective Effects of Probiotics against Alzheimer's Disease.
Mishra V, Yadav D, Solanki KS, Koul B, Song M. Mishra V, et al. Biology (Basel). 2023 Dec 22;13(1):8. doi: 10.3390/biology13010008. Biology (Basel). 2023. PMID: 38248439 Free PMC article. Review. - Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches.
Belizário JE, Napolitano M. Belizário JE, et al. Front Microbiol. 2015 Oct 6;6:1050. doi: 10.3389/fmicb.2015.01050. eCollection 2015. Front Microbiol. 2015. PMID: 26500616 Free PMC article. Review.
References
- Wooley JC, Godzik A, Friedberg I (2010) A primer on metagenomics. PLoS Comput Biol 6: e1000667 doi:10.1371/journal.pcbi.1000667. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- U54HG004969/HG/NHGRI NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01HG004885/HG/NHGRI NIH HHS/United States
- R01HG005975/HG/NHGRI NIH HHS/United States
- R01 HG005969/HG/NHGRI NIH HHS/United States
- U54 HG004969/HG/NHGRI NIH HHS/United States
- R01HG005969/HG/NHGRI NIH HHS/United States
- R01 HG005975/HG/NHGRI NIH HHS/United States
- R01 HG004885/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources