The labial gene is required to terminate proliferation of identified neuroblasts in postembryonic development of the Drosophila brain - PubMed (original) (raw)
The labial gene is required to terminate proliferation of identified neuroblasts in postembryonic development of the Drosophila brain
Philipp A Kuert et al. Biol Open. 2012.
Erratum in
- The labial gene is required to terminate proliferation of identified neuroblasts in postembryonic development of the Drosophila brain.
Kuert PA, Bello BC, Reichert H. Kuert PA, et al. Biol Open. 2016 Aug 15;5(8):1175. doi: 10.1242/bio.020230. Biol Open. 2016. PMID: 27412266 Free PMC article. No abstract available.
Abstract
The developing brain of Drosophila has become a useful model for studying the molecular genetic mechanisms that give rise to the complex neuronal arrays that characterize higher brains in other animals including mammals. Brain development in Drosophila begins during embryogenesis and continues during a subsequent postembryonic phase. During embryogenesis, the Hox gene labial is expressed in the developing tritocerebrum, and labial loss-of-function has been shown to be associated with a loss of regional neuronal identity and severe patterning defects in this part of the brain. However, nothing is known about the expression and function of labial, or any other Hox gene, during the postembryonic phase of brain development, when the majority of the neurons in the adult brain are generated. Here we report the first analysis of Hox gene action during postembryonic brain development in Drosophila. We show that labial is expressed initially in six larval brain neuroblasts, of which only four give rise to the labial expressing neuroblast lineages present in the late larval brain. Although MARCM-based clonal mutation of labial in these four neuroblast lineages does not result in an obvious phenotype, a striking and unexpected effect of clonal labial loss-of-function does occur during postembryonic brain development, namely the formation of two ectopic neuroblast lineages that are not present in wildtype brains. The same two ectopic neuroblast lineages are also observed following cell death blockage and, significantly, in this case the resulting ectopic lineages are Labial-positive. These findings imply that labial is required in two specific neuroblast lineages of the wildtype brain for the appropriate termination of proliferation through programmed cell death. Our analysis of labial function reveals a novel cell autonomous role of this Hox gene in shaping the lineage architecture of the brain during postembryonic development.
Keywords: Brain; Drosophila; Homeotic; Hox; Lineage; Neural stem cell; Neuroblast; Postembryonic; Programmed cell death; labial.
Conflict of interest statement
Competing interests: The authors have no competing interests to declare.
Figures
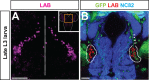
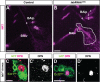
Fig. 1.. Regionalized expression of labial in the posterior central brain at the late larval stage.
(A) Overview of the late L3 larval brain. Two bilaterally symmetric cell clusters express labial. Labeled cells are shown in a Z-projection of multiple optical sections of a whole-mount brain. Dotted line indicates midline. Inset shows total larval CNS with box indicating region of labial expression. (B) Single optical section showing labial expressing cells (red), nc82 immunolabeled neuropile (blue) and MZ1407-Gal4 driven and membrane-targeted GFP expression (green). Dotted lines indicate position of the labial expressing cells. Arrowheads indicate secondary axon tract of labial expressing cells. Scale bars: 50 µm in A; 20 µm in B. In this and all subsequent figures, ventral views of the brain are presented and, with the exception of Fig. 3E–H′, anterior is always to the top.
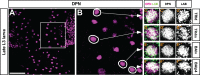
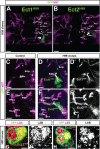
Fig. 2.. labial is expressed in four neuroblasts of the late L3 larval brain.
Labeled cells are shown in a Z-projection of multiple optical sections. (A) Overview of anti-Deadpan immunolabeled cells in the L3 larval brain. Box delimits cells a region in one hemisphere of the posterior central brain. (B) Magnified view of the boxed region shown in A. Neuroblasts co-immunolabeled with anti-Deadpan and anti-Labial are indicated by circles. (C–F″) Single optical sections of each of the four Deadpan-immunolabeled neuroblasts that express labial at the late L3 stage. Magnified view of the circled cells shown in B. Anti-Deadpan immunolabeling is in magenta. Labial immunolabeling is in green. Based on their relative position, each of these neuroblasts can be assigned to four lineages: TRld, TRdm, BAlv, BAlp4. Scale bar: 50 µm in A.
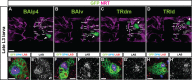
Fig. 3.. The four labial expressing neuroblasts give rise to the BAlp4, BAlv, TRdm and TRld lineage.
(A–D) Individual identified neuroblast clones are shown for each lineage together with the array of identified secondary axon tracts in the posterior central brain region of interest. GFP-labeled MARCM clones of the four neuroblast lineages are in green. Anti-Neurotactin labeling of secondary axon tracts secondary lineages is in magenta. Arrows indicate position of the cell bodies of the BAlp4, BAlv, TRdm and TRld lineages. Arrowheads indicate secondary axon tracts of the BAlp4, BAlv, TRdm and TRld lineages. Figures are superposition of multiple optical sections in late L3 brains. (E–H′) Neuroblasts in each of these four lineages express labial. Deadpan immunolabeling (neuroblasts) is in blue. Labial immunolabeling is in red. Single optical sections of BAlp4 (E,E′), BAlv (F,F′), TRdm (G,G′) and TRld (H,H′). Stars indicate the neuroblast. Scale bars: 20 µm in A–D; 5 µm in E–H′.
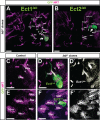
Fig. 4.. Clonal loss-of-function of labial leads to the formation of ectopic neuroblast lineages.
Late L3 brains; GFP-labeled mutant lab_14 MARCM clones are in green; secondary axon tracts labeled by anti-Neurotactin are in magenta. (A,B) Two identified ectopic neuroblast clones, Ect1_lab and Ect2_lab_ (arrows), are recovered in labial clonal loss-of-function experiments. Arrowheads indicate secondary axon tracts of Ect1_lab_ and Ect2_lab._ Figures are superposition of multiple optical sections. (C–F′) Both of these ectopic lineages can be identified by the projection patterns of their ectopic secondary axon tracts relative to the surrounding wildtype secondary axon tract scaffold. (C,E) Control showing corresponding wildtype axon tracts in two different optical sections. (D,D′) Ect1_lab_ is present between the labial expressing lineages BAlp4 and BAlv and projects several axon bundles in anterior-medial direction. Single optical section. (F,F′) Ect2_lab_ is close to the labial expressing lineages TRdm and TRld. Single optical section. Scale bars: 20 µm in A,B; 10 µm in C–F′.
Fig. 5.. Targeted RNAi knockdown of labial leads to ectopic neuroblast lineages comparable to those induced by labial loss-of-function mutation.
Late L3 brains. (A) Wildtype control showing the secondary axon tracts of the BAlp1–4 and BAlv lineages. Anti-NRT immunolabeling, Z-projection of optical sections. (B) UAS-lab_RNAi2990 driven by MZ1407-Gal4 to knockdown labial results in ectopic lineages. Dotted lines indicate position of Ect1_lab ectopic lineage relative to the secondary axon tracts of the BAlp1–4 and the BAlv lineages. Anti-NRT immunolabeling, Z-projections of optical sections. (C,D) Ectopic lineages contain a single Deadpan-positive neuroblast. GFP labeled lab_14 MARCM mutant clones of Ect1_lab and Ect2_lab_ (green) immunostained with anti-Deadpan (magenta). Single optical sections. Stars indicate ectopic neuroblasts. Scale bars: 10 µm in A–D′.
Fig. 6.. Six labial expressing neuroblasts are present at the L2 larval stage.
(A) Overview of anti-Deadpan immunolabeled cells in the late L2 larval brain. Labeled cells are shown in a Z-projection of multiple optical sections. (B) Magnified view of the region in the box of A. Neuroblasts co-immunolabeled with anti-Deadpan and anti-Labial are indicated by circles. (C–H″) Single optical sections of each of the six anti-Deadpan immunolabeled neuroblasts that express labial at the late L2 stage. Magnification of the circled cells shown in B. Based on their relative position and appearance, each of these neuroblasts can be assigned to the BAlp4, BAlv, TRdm, TRld, Ect1_lab_, and Ect2_lab_ lineages. Deadpan immunolabeling is in magenta. Labial immunolabeling is in green. (I) Overview of anti-Deadpan immunolabeled cells in the early L3 larval brain. (J) Magnified view of the region in the box of I. Neuroblasts co-immunolabeled with anti-Deadpan and anti-Labial are indicated by circles. (K–O″) Single optical sections of each of the four anti-Deadpan-immunolabeled neuroblasts that express labial at the early L3 stage. Magnification of the circled cells shown in J. Based on their relative position, each of these neuroblasts can be assigned to the BAlp4, BAlv, TRdm, and TRld lineages. Deadpan immunolabeling is in magenta. Labial immunolabeling is in green. Scale bars: 20 µm in A,I.
Fig. 7.. Blocking of cell death leads to labial expressing ectopic neuroblast lineages.
GFP-labeled H99 MARCM mutant clones are green; secondary axon tracts labeled by anti-Neurotactin are in magenta. (A,B) Two identified ectopic neuroblast clones, Ect1H99 and Ect2H99 (arrows) are recovered after cell death block; both are similar in terms of position and secondary axon projection pattern (arrowheads) to Ect1_lab_ and Ect2_lab_ found in the labial mutant assay. Superposition of multiple optical sections. (C,E) Control showing corresponding wildtype axon tracts in two different optical sections. (D,D′,F,F′) Following clonal cell death block, both ectopic lineages can be identified by the projection patterns of their ectopic secondary axon tracts relative to the surrounding wildtype secondary axon tract scaffold. Single optical sections. (G,H) The ectopic Ect1H99 and Ect2H99 lineages express labial in the neuroblast (star). Labial immunolabeling is in magenta. Scale bars: 20 µm in A,B; 10 µm in C–F′; 5 µm in G–H′.
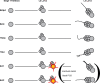
Fig. 8.. Model for _labial_-dependent termination of specific postembryonic neuroblasts.
In this model, labial is cell autonomously required for the stage-specific programmed cell death of two of the six postembryonic neuroblasts that express labial during larval stages. The neuroblasts of the BAlp4, BAlv, TRdm and TRld lineages express labial and persist to late L3 larval stages. The neuroblasts of the Ect1 and Ect2 lineage express labial during early larval stages, and labial is required for their termination through apoptosis at the L2 to L3 transition. If labial is inactivated or cell death is blocked, these two neuroblasts give rise to ectopic lineages which persist to late L3 larval stages.
Similar articles
- Neuroblast lineage identification and lineage-specific Hox gene action during postembryonic development of the subesophageal ganglion in the Drosophila central brain.
Kuert PA, Hartenstein V, Bello BC, Lovick JK, Reichert H. Kuert PA, et al. Dev Biol. 2014 Jun 15;390(2):102-15. doi: 10.1016/j.ydbio.2014.03.021. Epub 2014 Apr 5. Dev Biol. 2014. PMID: 24713419 - Polycomb group genes are required for neural stem cell survival in postembryonic neurogenesis of Drosophila.
Bello B, Holbro N, Reichert H. Bello B, et al. Development. 2007 Mar;134(6):1091-9. doi: 10.1242/dev.02793. Epub 2007 Feb 7. Development. 2007. PMID: 17287254 - Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain.
Izergina N, Balmer J, Bello B, Reichert H. Izergina N, et al. Neural Dev. 2009 Dec 11;4:44. doi: 10.1186/1749-8104-4-44. Neural Dev. 2009. PMID: 20003348 Free PMC article. - Control of neural stem cell self-renewal and differentiation in Drosophila.
Kang KH, Reichert H. Kang KH, et al. Cell Tissue Res. 2015 Jan;359(1):33-45. doi: 10.1007/s00441-014-1914-9. Epub 2014 Jun 6. Cell Tissue Res. 2015. PMID: 24902665 Review. - Embryonic development of the insect central complex: insights from lineages in the grasshopper and Drosophila.
Boyan G, Williams L. Boyan G, et al. Arthropod Struct Dev. 2011 Jul;40(4):334-48. doi: 10.1016/j.asd.2011.02.005. Epub 2011 Mar 5. Arthropod Struct Dev. 2011. PMID: 21382507 Review.
Cited by
- Hox targets and cellular functions.
Sánchez-Herrero E. Sánchez-Herrero E. Scientifica (Cairo). 2013;2013:738257. doi: 10.1155/2013/738257. Epub 2013 Dec 30. Scientifica (Cairo). 2013. PMID: 24490109 Free PMC article. - Anterior Hox Genes and the Process of Cephalization.
Hombría JC, García-Ferrés M, Sánchez-Higueras C. Hombría JC, et al. Front Cell Dev Biol. 2021 Aug 5;9:718175. doi: 10.3389/fcell.2021.718175. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34422836 Free PMC article. Review. - Clonal development and organization of the adult Drosophila central brain.
Yu HH, Awasaki T, Schroeder MD, Long F, Yang JS, He Y, Ding P, Kao JC, Wu GY, Peng H, Myers G, Lee T. Yu HH, et al. Curr Biol. 2013 Apr 22;23(8):633-43. doi: 10.1016/j.cub.2013.02.057. Epub 2013 Mar 28. Curr Biol. 2013. PMID: 23541733 Free PMC article. - Timelines in the insect brain: fates of identified neural stem cells generating the central complex in the grasshopper Schistocerca gregaria.
Boyan G, Liu Y. Boyan G, et al. Dev Genes Evol. 2014 Feb;224(1):37-51. doi: 10.1007/s00427-013-0462-8. Epub 2013 Dec 17. Dev Genes Evol. 2014. PMID: 24343526
References
- Abrams J. M., White K., Fessler L. I., Steller H. (1993). Programmed cell death during Drosophila embryogenesis. Development 117, 29–43. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases