Viral diversity threshold for adaptive immunity in prokaryotes - PubMed (original) (raw)
Viral diversity threshold for adaptive immunity in prokaryotes
Ariel D Weinberger et al. mBio. 2012.
Abstract
Bacteria and archaea face continual onslaughts of rapidly diversifying viruses and plasmids. Many prokaryotes maintain adaptive immune systems known as clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated genes (Cas). CRISPR-Cas systems are genomic sensors that serially acquire viral and plasmid DNA fragments (spacers) that are utilized to target and cleave matching viral and plasmid DNA in subsequent genomic invasions, offering critical immunological memory. Only 50% of sequenced bacteria possess CRISPR-Cas immunity, in contrast to over 90% of sequenced archaea. To probe why half of bacteria lack CRISPR-Cas immunity, we combined comparative genomics and mathematical modeling. Analysis of hundreds of diverse prokaryotic genomes shows that CRISPR-Cas systems are substantially more prevalent in thermophiles than in mesophiles. With sequenced bacteria disproportionately mesophilic and sequenced archaea mostly thermophilic, the presence of CRISPR-Cas appears to depend more on environmental temperature than on bacterial-archaeal taxonomy. Mutation rates are typically severalfold higher in mesophilic prokaryotes than in thermophilic prokaryotes. To quantitatively test whether accelerated viral mutation leads microbes to lose CRISPR-Cas systems, we developed a stochastic model of virus-CRISPR coevolution. The model competes CRISPR-Cas-positive (CRISPR-Cas+) prokaryotes against CRISPR-Cas-negative (CRISPR-Cas-) prokaryotes, continually weighing the antiviral benefits conferred by CRISPR-Cas immunity against its fitness costs. Tracking this cost-benefit analysis across parameter space reveals viral mutation rate thresholds beyond which CRISPR-Cas cannot provide sufficient immunity and is purged from host populations. These results offer a simple, testable viral diversity hypothesis to explain why mesophilic bacteria disproportionately lack CRISPR-Cas immunity. More generally, fundamental limits on the adaptability of biological sensors (Lamarckian evolution) are predicted.
Importance: A remarkable recent discovery in microbiology is that bacteria and archaea possess systems conferring immunological memory and adaptive immunity. Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated genes (CRISPR-Cas) are genomic sensors that allow prokaryotes to acquire DNA fragments from invading viruses and plasmids. Providing immunological memory, these stored fragments destroy matching DNA in future viral and plasmid invasions. CRISPR-Cas systems also provide adaptive immunity, keeping up with mutating viruses and plasmids by continually acquiring new DNA fragments. Surprisingly, less than 50% of mesophilic bacteria, in contrast to almost 90% of thermophilic bacteria and Archaea, maintain CRISPR-Cas immunity. Using mathematical modeling, we probe this dichotomy, showing how increased viral mutation rates can explain the reduced prevalence of CRISPR-Cas systems in mesophiles. Rapidly mutating viruses outrun CRISPR-Cas immune systems, likely decreasing their prevalence in bacterial populations. Thus, viral adaptability may select against, rather than for, immune adaptability in prokaryotes.
Figures
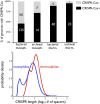
FIG 1
CRISPR-Cas is disproportionately prevalent in thermophiles. (Top) Bar graph showing the percentage of CRISPR-Cas+ and CRISPR-Cas− prokaryotes in mesophilic (mesoph.) and thermophilic (therm.) environments. The numbers in white or black shown on the bars are the numbers of species. Across 383 diversified bacterial and archaeal genomes (Materials and Methods), CRISPR-Cas is disproportionately sampled in thermophiles (P < 10−6). (Bottom) CRISPR locus length distributions. The distributions of locus lengths for CRISPR-Cas+ thermophiles and mesophiles fit to log10 of the total number of spacers per genome (Fig. S1 shows histograms in logarithmic and nonlogarithmic scales). On average, thermophilic loci possess more spacers (P < 10−7). However, CRISPR locus lengths show greater variance in mesophiles (P = 5 × 10−3).
FIG 2
Model of virus-host coevolution. Schematic of a representative model iteration. Host and viral populations are divided into strains labeled B1 and B2 (B stands for bacteria) and V1 and V2 (V stands for virus), respectively. Host strains are shown as ovals and viral strains as hexagons; bars within the strains reflect host spacers and viral protospacers. Identical spacer and protospacer sequences have the same colored bar. Host strains lacking CRISPR-Cas are shown as empty ovals. In the first step of a model iteration, host and viral strains encounter one another. In the next step, mutations occur (single and double quotation marks). Because viruses mutate only in productive encounters and hosts incorporate spacers only if they survive infection, mutations are concentrated in immune hosts and infective viruses. In the next step, more mutations can occur if CRISPR-Cas+ hosts delete spacers or lose entire CRISPR-Cas loci. Similarly, CRISPR-Cas− hosts can acquire CRISPR-Cas loci. In the fourth step, selection modifies strain frequencies according to equations 1 and 2 in the text. Finally, each iteration ends with the model randomly sampling approximately fixed numbers of hosts and viruses.
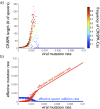
FIG 3
Rapid viral mutation overwhelms CRISPR-Cas systems. (a) Average CRISPR locus length (y axis) and prevalence (heatmap) as functions of the viral mutation rate. Model averages are “time averages” taken across 100,000 (10_N_) iterations. Viral mutation rates range from 10−4 to 10−2 per genome per productive infection. Since spacer deletion rates outpace spacer addition rates (see Table S1 in the supplemental material), evolution prunes CRISPR loci to the smallest sizes at which they provide antiviral immunity. At low viral mutation rates, few spacers are required against the highly similar viruses. As the viral mutation rate increases, locus lengths increase, with more spacers required against the diversifying viruses (see Fig. S3 in the supplemental material). Once viral mutation rates increase above a threshold, however, CRISPR-Cas systems can no longer keep pace. Overwhelmed by viral diversity, locus lengths crash to 0, and the system is purged from host populations. (b) Effective host and viral mutation rates as functions of the viral mutation rate. Since viruses mutate protospacers only during productive virus-host encounters, the effective viral mutation rate in a simulation is the product of the fraction of productive encounters and the (changing) viral mutation rate. Similarly, since hosts add spacers only during immune virus-host encounters, the effective host mutation rate in a simulation is the product of the fraction of immune encounters and the (fixed) spacer addition rate. Notably, the longest CRISPR loci emerge at reduced effective spacer addition rates. In fact, these long loci emerge in the small window in which the effective mutation rates of both host and virus are nonnegligible. Locus lengths thus reflect the strength of virus-host coevolution, rather than the rate of host spacer addition.
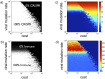
FIG 4
Cost-benefit analysis of CRISPR-Cas prevalence. Heatmaps of model statistics—averaged over 100,000 iterations—as functions of the cost of CRISPR-Cas and the viral mutation rate. (a) CRISPR-Cas prevalence. Both high costs and high viral mutation rates shift host populations from 100% CRISPR-Cas+ to 0% CRISPR-Cas+. Importantly, the intermediate CRISPR-Cas frequencies found in the midregion separating these two extremes (i.e., the separatrix) do not reflect coexisting CRISPR-Cas+ and CRISPR-Cas− populations. Instead, the separatrix frequencies reflect an average over time as the model alternates between quasi-steady states of entirely CRISPR-Cas+ and entirely CRISPR-Cas− populations (see Fig. S4 and Fig. S5 in the supplemental material). (b) Probability that CRISPR-Cas provides immunity. Given the low failure rate of CRISPR-Cas (Table S1), the probability of CRISPR-Cas providing immunity is effectively the probability that a host spacer matches a viral protospacer. High viral mutation rates thus reduce CRISPR-Cas immunity. In fact, at high viral mutation rates, immunity is absent even when CRISPR-Cas remains 100% prevalent due to low costs (top left of figure 4b). (c) Average (Shannon) diversity of viral protospacers. Increasing the viral mutation rate increases the average diversity across time by increasing genetic diversity at each time point. More surprisingly, increasing the cost of CRISPR-Cas above a threshold rapidly increases viral diversity, because it purges CRISPR-Cas from populations, allowing viruses to freely mutate. (d) Average CRISPR-Cas locus lengths. As in Fig. 3a, which reflects a vertical cross section of Fig. 5d (i.e., locus lengths at a single cost), the largest CRISPR loci emerge at intermediate viral mutation rates.
Similar articles
- Pseudo-chaotic oscillations in CRISPR-virus coevolution predicted by bifurcation analysis.
Berezovskaya FS, Wolf YI, Koonin EV, Karev GP. Berezovskaya FS, et al. Biol Direct. 2014 Jul 2;9:13. doi: 10.1186/1745-6150-9-13. Biol Direct. 2014. PMID: 24986220 Free PMC article. - CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes.
Koonin EV, Makarova KS. Koonin EV, et al. RNA Biol. 2013 May;10(5):679-86. doi: 10.4161/rna.24022. Epub 2013 Feb 25. RNA Biol. 2013. PMID: 23439366 Free PMC article. Review. - Evolutionary dynamics of the prokaryotic adaptive immunity system CRISPR-Cas in an explicit ecological context.
Iranzo J, Lobkovsky AE, Wolf YI, Koonin EV. Iranzo J, et al. J Bacteriol. 2013 Sep;195(17):3834-44. doi: 10.1128/JB.00412-13. Epub 2013 Jun 21. J Bacteriol. 2013. PMID: 23794616 Free PMC article. - CRISPR-Cas systems and RNA-guided interference.
Barrangou R. Barrangou R. Wiley Interdiscip Rev RNA. 2013 May-Jun;4(3):267-78. doi: 10.1002/wrna.1159. Epub 2013 Mar 20. Wiley Interdiscip Rev RNA. 2013. PMID: 23520078 Review. - Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes.
Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Al-Attar S, et al. Biol Chem. 2011 Apr;392(4):277-89. doi: 10.1515/BC.2011.042. Epub 2011 Feb 7. Biol Chem. 2011. PMID: 21294681 Review.
Cited by
- Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex.
Blosser TR, Loeff L, Westra ER, Vlot M, Künne T, Sobota M, Dekker C, Brouns SJJ, Joo C. Blosser TR, et al. Mol Cell. 2015 Apr 2;58(1):60-70. doi: 10.1016/j.molcel.2015.01.028. Epub 2015 Mar 5. Mol Cell. 2015. PMID: 25752578 Free PMC article. - Life in hot acid: a genome-based reassessment of the archaeal order Sulfolobales.
Counts JA, Willard DJ, Kelly RM. Counts JA, et al. Environ Microbiol. 2021 Jul;23(7):3568-3584. doi: 10.1111/1462-2920.15189. Epub 2020 Sep 7. Environ Microbiol. 2021. PMID: 32776389 Free PMC article. - Precipitous Increase of Bacterial CRISPR-Cas Abundance at Around 45°C.
Lan XR, Liu ZL, Niu DK. Lan XR, et al. Front Microbiol. 2022 Mar 1;13:773114. doi: 10.3389/fmicb.2022.773114. eCollection 2022. Front Microbiol. 2022. PMID: 35300480 Free PMC article. - Ecological memory preserves phage resistance mechanisms in bacteria.
Skanata A, Kussell E. Skanata A, et al. Nat Commun. 2021 Nov 24;12(1):6817. doi: 10.1038/s41467-021-26609-w. Nat Commun. 2021. PMID: 34819498 Free PMC article. - Addiction systems antagonize bacterial adaptive immunity.
van Sluijs L, van Houte S, van der Oost J, Brouns SJ, Buckling A, Westra ER. van Sluijs L, et al. FEMS Microbiol Lett. 2019 Mar 1;366(5):fnz047. doi: 10.1093/femsle/fnz047. FEMS Microbiol Lett. 2019. PMID: 30834930 Free PMC article.
References
- Futuyma D. 2005. Evolution. Sinauer Associates, Sunderland, MA
- Lynch M. 2007. The origins of genome architecture. Sinauer Associates, Sunderland, MA
- Eyre-Walker A, Keightley PD. 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8:610–618 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources