Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines - PubMed (original) (raw)
. 2012 Dec 11;3(6):e00515-12.
doi: 10.1128/mBio.00515-12.
V Stalin Raj, Doreen Muth, Benjamin Meyer, Stephan Kallies, Saskia L Smits, Robert Wollny, Theo M Bestebroer, Sabine Specht, Tasnim Suliman, Katrin Zimmermann, Tabea Binger, Isabella Eckerle, Marco Tschapka, Ali M Zaki, Albert D M E Osterhaus, Ron A M Fouchier, Bart L Haagmans, Christian Drosten
Affiliations
- PMID: 23232719
- PMCID: PMC3520110
- DOI: 10.1128/mBio.00515-12
Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines
Marcel A Müller et al. mBio. 2012.
Abstract
A new human coronavirus (hCoV-EMC) has emerged very recently in the Middle East. The clinical presentation resembled that of the severe acute respiratory syndrome (SARS) as encountered during the epidemic in 2002/2003. In both cases, acute renal failure was observed in humans. HCoV-EMC is a member of the same virus genus as SARS-CoV but constitutes a sister species. Here we investigated whether it might utilize angiotensin-converting enzyme 2 (ACE2), the SARS-CoV receptor. Knowledge of the receptor is highly critical because the restriction of the SARS receptor to deep compartments of the human respiratory tract limited the spread of SARS. In baby hamster kidney (BHK) cells, lentiviral transduction of human ACE2 (hACE2) conferred permissiveness and replication for SARS-CoV but not for hCoV-EMC. Monkey and human kidney cells (LLC-MK2, Vero, and 769-P) and swine kidney cells were permissive for both viruses, but only SARS-CoV infection could be blocked by anti-hACE2 antibody and could be neutralized by preincubation of virus with soluble ACE2. Our data show that ACE2 is neither necessary nor sufficient for hCoV-EMC replication. Moreover, hCoV-EMC, but not SARS-CoV, replicated in cell lines from Rousettus, Rhinolophus, Pipistrellus, Myotis, and Carollia bats, representing four major chiropteran families from both suborders. As human CoV normally cannot replicate in bat cells from different families, this suggests that hCoV-EMC might use a receptor molecule that is conserved in bats, pigs, and humans, implicating a low barrier against cross-host transmission. IMPORTANCE A new human coronavirus (hCoV) emerged recently in the Middle East. The disease resembled SARS (severe acute respiratory syndrome), causing a fatal epidemic in 2002/2003. Coronaviruses have a reservoir in bats and because this novel virus is related to SARS-CoV, we investigated whether it might replicate in bat cells and use the same receptor (angiotensin-converting enzyme 2 [ACE2]). This knowledge is highly critical, because the SARS-CoV receptor influenced pathology, and its localization in the deep respiratory tract is thought to have restricted the transmissibility of SARS. Our data show that hCoV-EMC does not need the SARS-CoV receptor to infect human cells. Moreover, the virus is capable of infecting human, pig, and bat cells. This is remarkable, as human CoVs normally cannot replicate in bat cells as a consequence of host adaptation. Our results implicate that the new virus might use a receptor that is conserved between bats, pigs and humans suggesting a low barrier against cross-host transmission.
Figures
FIG 1
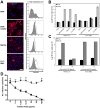
Human ACE2-independent entry of hCoV-EMC. (A) Baby hamster kidney (BHK) cells were transduced with hACE2-carrying lentiviruses and selected by puromycin treatment. (Left) hACE2 expression was controlled by immunofluorescence assay (IFA) by using goat anti-hACE2 immunoglobulin (Ig) (R&D Systems) followed by cyanin 3-labeled donkey anti-goat Ig (Dianova). For controls, ACE2-expressing primate cell lines (kidney cells from rhesus monkey [LLC-MK2] and African green monkey [MA104]) were stained in parallel. DAPI, 4',6'-diamidino-2-phenylindole. Bar, 50 µm. (Right) ACE2 protein surface expression was determined by FACS analysis using a mouse anti-hACE2 Ig antibody (R&D Systems) in combination with an Alexa Fluor 647-labeled goat anti-mouse Ig (Life Technologies). Data were acquired using FACS Canto and analyzed with FlowJo software. The cells were incubated with the secondary antibody only (shown in gray) for a control. mAB, monoclonal antibody. (B) The interferon-deficient BHK cells and the generated hACE2-expressing transgenic BHK cells were applied in infection studies. LLC-MK2 and MA104 primate cells known to be susceptible for SARS-CoV were infected in parallel with SARS-CoV strain Frankfurt-1 and hCoV-EMC. The cells were infected with an MOI of 0.5 for 1 h, washed twice with phosphate-buffered saline (PBS), and supplemented with medium. At time points 0 and 40 h after infection, samples from supernatants were taken in order to extract viral RNA. Real-time reverse transcription-PCR (RT-PCR) was used for absolute quantification of genome equivalents (GE) per ml of supernatant (2, 10). All experiments were performed in triplicate. hpi, hours postinfection. (C) In order to block the ACE2 receptor, Vero cells were preincubated with 10 µg/ml of polyclonal goat anti-hACE2 for 1 h at 37°C. Half of the antibody solution was stored, and cells were subsequently infected by adding viruses at an MOI of 0.01 for 30 min at 4°C. The supernatant was discarded, and medium with antibody solution was added. At time points 0 and 20 h postinfection, samples from supernatants (triplicate samples) were taken for quantification by real-time RT-PCR. pAb, polyclonal antibody. (D) Virus-neutralizing activity of soluble recombinant hACE2 (rhACE2) was tested by preincubating in triplicate different concentrations of rhACE2 (a kind gift from J. M. Penninger, Vienna, Austria) with 104 50% tissue culture infective dose (TCID50)/ml hCoV-EMC (triangles) or 103 TCID50/ml SARS-CoV (squares) for 1 h. The mixture was added to Vero E6 cells for 1 h, after which cells were washed and fresh medium was added. The infection was stopped after 8 h, and cells were visualized with a cross-reactive SARS-CoV serum followed by a goat anti-rabbit IgG antibody conjugated to peroxidase and stained with 3-amino-9-ethylcarbazole (AEC) substrate.
FIG 2
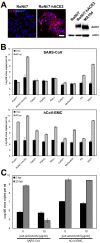
Bat cells allow ACE2-independent entry of hCoV-EMC. (A) Immortalized Rousettus aegyptiacus cells were provided with hACE2. (Left) Expression of hACE2 was controlled by immunofluorescence assay using goat anti-hACE2 Ig and cyanin 3-labeled donkey anti-goat Ig. Bar, 50 µm. (Right) Western blot analysis was done using goat anti-hACE2 Ig followed by horseradish peroxidase-labeled donkey anti-goat Ig (Dianova). For a control, a protein lysate of MA104 cells was applied to the blot. A loading control was done with a rabbit anti-pan-actin Ig (Cell Signaling). (B) Representative bat cell cultures from Yinpterochiroptera (Rousettus aegyptiacus kidney [RoNi/7] and Rhinolophus landeri lung [RhiLu]) and Yangochiroptera (Pipistrellus pipistrellus kidney [PipNi/1 and PipNi/3], Carollia perspicillata kidney [CarNi/1], and Myotis daubentonii kidney [MyDauNi/2]) and porcine (PS) and human kidney (769-P) cell lines were used for infection studies with SARS-CoV (top panel) and hCoV-EMC (bottom panel). For a control and for comparison, hACE2-expressing transgenic Rousettus bat cell cultures (RoNi/7-hACE2) were included. The cells were infected at an MOI of 0.5 for 1 h, washed twice with PBS, and supplemented with medium. At time points 0 and 40 h postinfection, samples from supernatants were taken to extract viral RNA. Real-time RT-PCR was performed as described for absolute quantification of GE. All experiments were performed in triplicate. (C) A hACE2-blocking experiment was done with RoNi/7-hACE2 cells. The cells were preincubated with 10 µg/ml of polyclonal goat anti-hACE2 for 1 h at 37°C. The cells were subsequently infected by adding viruses at an MOI of 0.01 for 30 min at 4°C. The supernatant was discarded, and medium supplemented with hACE2 antibody was added. At time points 0 and 20 h postinfection, samples from supernatants were taken to quantify GE by real-time RT-PCR. The experiment was performed in triplicate.
Comment in
- Human coronavirus EMC is not the same as severe acute respiratory syndrome coronavirus.
Perlman S, Zhao J. Perlman S, et al. mBio. 2013 Jan 15;4(1):e00002-13. doi: 10.1128/mBio.00002-13. mBio. 2013. PMID: 23322635 Free PMC article.
Similar articles
- Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential.
Kindler E, Jónsdóttir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, Geffers R, Fouchier RA, Drosten C, Müller MA, Dijkman R, Thiel V. Kindler E, et al. mBio. 2013 Feb 19;4(1):e00611-12. doi: 10.1128/mBio.00611-12. mBio. 2013. PMID: 23422412 Free PMC article. - Human coronavirus EMC is not the same as severe acute respiratory syndrome coronavirus.
Perlman S, Zhao J. Perlman S, et al. mBio. 2013 Jan 15;4(1):e00002-13. doi: 10.1128/mBio.00002-13. mBio. 2013. PMID: 23322635 Free PMC article. - Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts.
Zhai X, Sun J, Yan Z, Zhang J, Zhao J, Zhao Z, Gao Q, He WT, Veit M, Su S. Zhai X, et al. J Virol. 2020 Jul 16;94(15):e00831-20. doi: 10.1128/JVI.00831-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32404529 Free PMC article. - Molecular mechanisms of human coronavirus NL63 infection and replication.
Castillo G, Mora-Díaz JC, Breuer M, Singh P, Nelli RK, Giménez-Lirola LG. Castillo G, et al. Virus Res. 2023 Apr 2;327:199078. doi: 10.1016/j.virusres.2023.199078. Epub 2023 Feb 22. Virus Res. 2023. PMID: 36813239 Free PMC article. Review. - Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.
Drexler JF, Corman VM, Drosten C. Drexler JF, et al. Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review.
Cited by
- Putative conformations of the receptor-binding domain in S protein of hCoV-EMC in complex with its receptor dipeptidyl peptidase-4.
Jiang S, Lu L, Du L, Debnath AK. Jiang S, et al. J Infect. 2013 Aug;67(2):156-8. doi: 10.1016/j.jinf.2013.04.007. Epub 2013 Apr 17. J Infect. 2013. PMID: 23603488 Free PMC article. No abstract available. - Middle East respiratory syndrome coronavirus (MERS-CoV): A review.
Ramadan N, Shaib H. Ramadan N, et al. Germs. 2019 Mar 1;9(1):35-42. doi: 10.18683/germs.2019.1155. eCollection 2019 Mar. Germs. 2019. PMID: 31119115 Free PMC article. Review. - Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation.
Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, Godeke GJ, Jonges M, Farag E, Diab A, Ghobashy H, Alhajri F, Al-Thani M, Al-Marri SA, Al Romaihi HE, Al Khal A, Bermingham A, Osterhaus AD, AlHajri MM, Koopmans MP. Haagmans BL, et al. Lancet Infect Dis. 2014 Feb;14(2):140-5. doi: 10.1016/S1473-3099(13)70690-X. Epub 2013 Dec 17. Lancet Infect Dis. 2014. PMID: 24355866 Free PMC article. - Bat airway epithelial cells: a novel tool for the study of zoonotic viruses.
Eckerle I, Ehlen L, Kallies R, Wollny R, Corman VM, Cottontail VM, Tschapka M, Oppong S, Drosten C, Müller MA. Eckerle I, et al. PLoS One. 2014 Jan 13;9(1):e84679. doi: 10.1371/journal.pone.0084679. eCollection 2014. PLoS One. 2014. PMID: 24454736 Free PMC article.
References
- Saif LJ. 2004. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev. Sci. Tech. 23:643–660 - PubMed
- Drosten C, et al. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967–1976 - PubMed
- Peiris JS, Yuen KY, Osterhaus AD, Stöhr K. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431–2441 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous