Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model - PubMed (original) (raw)
Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model
Pieter Van den Abbeele et al. ISME J. 2013 May.
Abstract
The human gut is colonized by a complex microbiota with multiple benefits. Although the surface-attached, mucosal microbiota has a unique composition and potential to influence human health, it remains difficult to study in vivo. Therefore, we performed an in-depth microbial characterization (human intestinal tract chip (HITChip)) of a recently developed dynamic in vitro gut model, which simulates both luminal and mucosal gut microbes (mucosal-simulator of human intestinal microbial ecosystem (M-SHIME)). Inter-individual differences among human subjects were confirmed and microbial patterns unique for each individual were preserved in vitro. Furthermore, in correspondence with in vivo studies, Bacteroidetes and Proteobacteria were enriched in the luminal content while Firmicutes rather colonized the mucin layer, with Clostridium cluster XIVa accounting for almost 60% of the mucin-adhered microbiota. Of the many acetate and/or lactate-converting butyrate producers within this cluster, Roseburia intestinalis and Eubacterium rectale most specifically colonized mucins. These 16S rRNA gene-based results were confirmed at a functional level as butyryl-CoA:acetate-CoA transferase gene sequences belonged to different species in the luminal as opposed to the mucin-adhered microbiota, with Roseburia species governing the mucosal butyrate production. Correspondingly, the simulated mucosal environment induced a shift from acetate towards butyrate. As not only inter-individual differences were preserved but also because compared with conventional models, washout of relevant mucin-adhered microbes was avoided, simulating the mucosal gut microbiota represents a breakthrough in modeling and mechanistically studying the human intestinal microbiome in health and disease. Finally, as mucosal butyrate producers produce butyrate close to the epithelium, they may enhance butyrate bioavailability, which could be useful in treating diseases, such as inflammatory bowel disease.
Figures
Figure 1
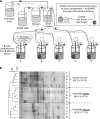
(a) Schematic representation of the main/first experiment: five colon compartments with a simulated mucosal environment (M-SHIME) were each inoculated with a different human faecal sample (donor A, B, C, D or E). (b) Dendrogram for the total bacterial DGGE profiles of the luminal and mucosal microbiota of these five M-SHIMEs, 3 days after inoculation with faecal samples of five human donors. The average inter-individual similarity between different donors (±s.e.m.) is shown for each type of sample: inoculum, lumen and mucin layer.
Figure 2
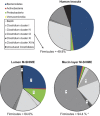
The average abundance (%) of higher taxonomic groups (∼ phylum level) based on the HITChip analysis of the human faecal inocula and the resulting luminal and mucosal environment of the M-SHIME, 3 days after inoculation (_n_=5). An asterisks indicates a significant difference in the abundance between the lumen and mucin layer of the M-SHIME. The averages and s.e. can be found in Supplementary Table S3.
Figure 3
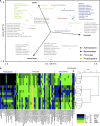
(a) Redundancy analysis at genus-like group-level based on the HITChip data of the inoculum, lumen and mucin layer of M-SHIMEs inoculated with faecal samples of five human subjects (_n_=5; _P_=0.02). (b) Dendrogram of the same samples based on the HITChip data for 131 genus-like groups, including a heat map which shows the inter-individual distribution of microbial groups that differed among inocula, lumen and/or mucin layer.
Figure 4
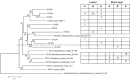
Phylogenetic tree and the amount of butyryl-CoA:acetate-CoA transferase gene sequences (∼480 bp) identified in the luminal content and mucin layer of the M-SHIME 68 h after inoculation with human faecal samples of different donors (A, B and D; first experiment). Sequences with at least 98% sequence identity were grouped together under the same OTU number of which one sequence was chosen to build the phylogenetic tree. OTU numbers >32 indicate novel butyrate-producing species identified during the current study.
Figure 5
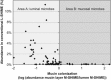
Average abundance (%) of genus-like groups as determined with the HITChip in a conventional L-SHIME without simulation of the mucosal environment, as reported previously (Van den Abbeele et al., 2010) (_n_=4), as a function of the mucin colonization of these microbes in the novel M-SHIME (_n_=5). Mucin colonization represents the preference of a bacterial group to colonize the in vitro mucosal environment and is expressed as the logarithm of the abundance in the mucosal environment versus the abundance in the luminal content of the M-SHIME. A value <0 indicates that the microbial group rather colonizes the lumen (area A), a value >0 indicates that this group rather colonizes the simulated mucosal environment (area B).
Similar articles
- Decreased colonization of fecal Clostridium coccoides/Eubacterium rectale species from ulcerative colitis patients in an in vitro dynamic gut model with mucin environment.
Vermeiren J, Van den Abbeele P, Laukens D, Vigsnaes LK, De Vos M, Boon N, Van de Wiele T. Vermeiren J, et al. FEMS Microbiol Ecol. 2012 Mar;79(3):685-96. doi: 10.1111/j.1574-6941.2011.01252.x. Epub 2011 Dec 2. FEMS Microbiol Ecol. 2012. PMID: 22092917 - Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats.
Van den Abbeele P, Gérard P, Rabot S, Bruneau A, El Aidy S, Derrien M, Kleerebezem M, Zoetendal EG, Smidt H, Verstraete W, Van de Wiele T, Possemiers S. Van den Abbeele P, et al. Environ Microbiol. 2011 Oct;13(10):2667-80. doi: 10.1111/j.1462-2920.2011.02533.x. Epub 2011 Aug 30. Environ Microbiol. 2011. PMID: 21883787 - Quantification of butyryl CoA:acetate CoA-transferase genes reveals different butyrate production capacity in individuals according to diet and age.
Hippe B, Zwielehner J, Liszt K, Lassl C, Unger F, Haslberger AG. Hippe B, et al. FEMS Microbiol Lett. 2011 Mar;316(2):130-5. doi: 10.1111/j.1574-6968.2010.02197.x. Epub 2011 Jan 17. FEMS Microbiol Lett. 2011. PMID: 21204931 - Understanding the effects of diet on bacterial metabolism in the large intestine.
Louis P, Scott KP, Duncan SH, Flint HJ. Louis P, et al. J Appl Microbiol. 2007 May;102(5):1197-208. doi: 10.1111/j.1365-2672.2007.03322.x. J Appl Microbiol. 2007. PMID: 17448155 Review. - Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine.
Louis P, Flint HJ. Louis P, et al. FEMS Microbiol Lett. 2009 May;294(1):1-8. doi: 10.1111/j.1574-6968.2009.01514.x. Epub 2009 Feb 13. FEMS Microbiol Lett. 2009. PMID: 19222573 Review.
Cited by
- Identification of mucin degraders of the human gut microbiota.
Raimondi S, Musmeci E, Candeliere F, Amaretti A, Rossi M. Raimondi S, et al. Sci Rep. 2021 May 27;11(1):11094. doi: 10.1038/s41598-021-90553-4. Sci Rep. 2021. PMID: 34045537 Free PMC article. - Impact of Western Diet on Enterohemorrhagic Escherichia coli Colonization in the Human In Vitro Mucosal Artificial Colon as Mediated by Gut Microbiota.
O'Sullivan D, Arora T, Durif C, Uriot O, Brun M, Riu M, Foguet-Romero E, Samarra I, Domingo-Almenara X, Gahan CGM, Etienne-Mesmin L, Blanquet-Diot S. O'Sullivan D, et al. Nutrients. 2024 Jun 27;16(13):2046. doi: 10.3390/nu16132046. Nutrients. 2024. PMID: 38999794 Free PMC article. - Impact of cystic fibrosis disease on archaea and bacteria composition of gut microbiota.
Miragoli F, Federici S, Ferrari S, Minuti A, Rebecchi A, Bruzzese E, Buccigrossi V, Guarino A, Callegari ML. Miragoli F, et al. FEMS Microbiol Ecol. 2017 Feb;93(2):fiw230. doi: 10.1093/femsec/fiw230. Epub 2016 Nov 2. FEMS Microbiol Ecol. 2017. PMID: 27810876 Free PMC article. - Capacity of a Microbial Synbiotic To Rescue the In Vitro Metabolic Activity of the Gut Microbiome following Perturbation with Alcohol or Antibiotics.
Tierney BT, Van den Abbeele P, Al-Ghalith GA, Verstrepen L, Ghyselinck J, Calatayud M, Marzorati M, Gadir AA, Daisley B, Reid G, Bron PA, Gevers D, Dhir R, Simmons SL. Tierney BT, et al. Appl Environ Microbiol. 2023 Mar 29;89(3):e0188022. doi: 10.1128/aem.01880-22. Epub 2023 Feb 22. Appl Environ Microbiol. 2023. PMID: 36840551 Free PMC article. - Age-Dependent Prebiotic Effects of Soluble Corn Fiber in M-SHIME® Gut Microbial Ecosystems.
Arroyo MC, Laurie I, Rotsaert C, Marzorati M, Risso D, Karnik K. Arroyo MC, et al. Plant Foods Hum Nutr. 2023 Mar;78(1):213-220. doi: 10.1007/s11130-023-01043-z. Epub 2023 Jan 25. Plant Foods Hum Nutr. 2023. PMID: 36694053 Free PMC article.
References
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials