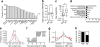Identification and analysis of murine pancreatic islet enhancers - PubMed (original) (raw)
doi: 10.1007/s00125-012-2797-5. Epub 2012 Dec 14.
A G Robertson, M Kramer, L Li, X Zhang, M Beach, N Thiessen, R Chiu, K Mungall, C J Whiting, P V Sabatini, A Kim, R Gottardo, M A Marra, F C Lynn, S J M Jones, P A Hoodless, B G Hoffman
Affiliations
- PMID: 23238790
- PMCID: PMC4773896
- DOI: 10.1007/s00125-012-2797-5
Identification and analysis of murine pancreatic islet enhancers
B R Tennant et al. Diabetologia. 2013 Mar.
Abstract
Aims/hypothesis: The paucity of information on the epigenetic barriers that are blocking reprogramming protocols, and on what makes a beta cell unique, has hampered efforts to develop novel beta cell sources. Here, we aimed to identify enhancers in pancreatic islets, to understand their developmental ontologies, and to identify enhancers unique to islets to increase our understanding of islet-specific gene expression.
Methods: We combined H3K4me1-based nucleosome predictions with pancreatic and duodenal homeobox 1 (PDX1), neurogenic differentiation 1 (NEUROD1), v-Maf musculoaponeurotic fibrosarcoma oncogene family, protein A (MAFA) and forkhead box A2 (FOXA2) occupancy data to identify enhancers in mouse islets.
Results: We identified 22,223 putative enhancer loci in in vivo mouse islets. Our validation experiments suggest that nearly half of these loci are active in regulating islet gene expression, with the remaining regions probably poised for activity. We showed that these loci have at least nine developmental ontologies, and that islet enhancers predominately acquire H3K4me1 during differentiation. We next discriminated 1,799 enhancers unique to islets and showed that these islet-specific enhancers have reduced association with annotated genes, and identified a subset that are instead associated with novel islet-specific long non-coding RNAs (lncRNAs).
Conclusions/interpretations: Our results indicate that genes with islet-specific expression and function tend to have enhancers devoid of histone methylation marks or, less often, that are bivalent or repressed, in embryonic stem cells and liver. Further, we identify a subset of enhancers unique to islets that are associated with novel islet-specific genes and lncRNAs. We anticipate that these data will facilitate the development of novel sources of functional beta cell mass.
Conflict of interest statement
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Figures
Fig. 1
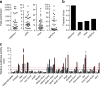
Identified loci have characteristics of functional enhancers. (a) Fold enrichment and (b) fraction of putative enhancer loci enriched for the indicated histone modifications and for p300 recruitment. The horizontal grey lines in (a) indicate the minimum fold enrichment needed for a locus to be considered enriched. (c) Relative luciferase activity levels (arbitrary units [AU]) of selected enhancer loci in HEK293 (white bars), Hepa1-6 (purple bars), mPAC (green bars), _α_TC-1 (blue bars) and MIN6 (red bars) cells. The black dashed line represents the mean relative luciferase activity of negative control regions in the cell lines, while the red dashed line indicates 2SD above this mean
Fig. 2
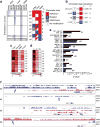
Identified putative enhancers in islets have distinct developmental ontologies. (a) Heatmap of the total read counts in ±1 kb regions around enhancer midpoints for H3K4me1, H3K4me3, H3K9me3 and H3K27me3 in ESCs, liver and islets. The data are segregated into enhancer clusters (e1–e9), and the chromatin state for each cluster is indicated in the schematic to the right of the heatmap. Squares composed of two triangles of different colours for clusters e5, e6, e8 and e9 indicate that these clusters have a mixed chromatin state. (b) The number and percentage of chromatin state transitions for the indicated enhancer clusters from ESCs to islets. (c, d) Heatmap of the fraction of enhancers in each enhancer cluster (c) occupied by PDX1, MAFA, NEUROD1 and FOXA2, or (d) single-, double-, triple- or quadruple-bound. (e) Enrichment p values of representative position weight matrices (PWMs) in the indicated enhancer clusters. (f) UCSC genome browser views of representative regions in the indicated enhancer clusters. H3K4me1 enrichment data are shown in blue, and H3K27me3 data in purple. All tracks are set to show a coverage depth range of 0 to 30. The arrows demarcate the identified putative enhancer loci
Fig. 3
Association of enhancer clusters with genes. (a) Box–whisker plot of expression levels in ESCs (light blue), liver (dark blue) and islets (red) of genes with enhancers in the indicated enhancer clusters. RPKM, reads per kilobase per million mapped reads. (b) Box–whisker plot of the specificity to islets as calculated using 203 SAGE libraries from different mouse tissues [34, 35] for genes associated with enhancers in each enhancer cluster. The red dashed line indicates the median for all expressed genes. (c) Heatmap of the enrichment of genes with low, intermediate or high specificity to islets (S) associated with enhancers in each cluster, relative to random expectation. Darker blue is more deprived, while darker red is more enriched. (d) Fold enrichment of significantly enriched GO and KEGG terms for genes with enhancers in the indicated enhancer clusters. Statistically significant differences in (a) and (b) were detected using a Kruskal–Wallis non-parametric test with a Dunn’s multiple comparison correction; *p<0.05, **p<0.01, ***p<0.001
Fig. 4
Identification of islet-specific putative enhancers. (a) The fraction of identified enhancer loci with H3K4me1 enrichment in 19 different mouse tissues or cell-types. ESC (129), ESCs from 129JAEC57/B6 mice; L1 day4 adi, 3T3-L1 cells 4 days after induction of adipogenesis; Liverf, Liver from 4f2a mice; L1 day2 adi, 3T3-L1 cells 2 days after induction of adipogenesis; Mefs, mouse embryonic fibroblasts; ESC (c57), ESCs from C57/B6 inbred mice; L1 day0 adi, 3T3-L1 cells on the day of induction of adipogenesis; Bmdm, bone marrow derived macrophages; Thiomacs, thioglycollate-elicited peritoneal macrophages; L1 pre-adi, 3T3-L1 cells 2 days before induction of adipogenesis; iPS, induced pluripotent stem cells; Bcells, resting splenic B cells; Neural prog, ESC-derived neural progenitors. Box–whisker plots of (b) expression level in islets (RNA-seq) and (c) specificity (SAGE) to islets of genes with associated ISEs or NSEs. The dashed line indicates the median of all expressed genes. Statistically significant differences were detected using a Kruskal–Wallis non-parametric test with a Dunn’s multiple comparison correction; ***p<0.001. RPKM, reads per kilobase per million mapped reads. (d) Fold enrichment of significantly enriched GO and KEGG terms for genes associated with ISE (white bars) and NSE (black bars) loci. (e) Frequency distribution of the GC content of DNA from ISEs (red) and NSEs (green). ****p<0.0001. (f) Fold enrichment of transcription factor binding motifs in ISEs relative to NSEs. (g) Distributions of the distance to the closest UCSC-known gene TSS within 200 kb for ISEs (red), NSEs (green) and random DNA (blue). (h) Fraction of loci that mapped (black) to Ensembl NCBMI.37 annotated transcripts or were not mappable (white) for ISEs and NSEs
Fig. 5
ISEs are associated with novel islet-specific lncRNAs. (a) UCSC mm9 genome browser views of representative novel transcripts with associated ISEs. The islet RNA-seq data from both replicates is overlaid in the first track below the novel transcripts. Below this is a track showing the transcription factor (TF) ChIP-seq data for PDX1 (purple), MAFA (red), NEUROD1 (green) and FOXA2 (yellow). The histone modification (HM) ChIP-seq data for H3K4me1 (purple) and H3K27me3 (red) is overlaid in the bottom track. Because no UCSC, Ensembl or Refseq genes are present within the regions, these tracks are not displayed. (b) Relative expression of identified lncRNAs with an associated ISE in islets compared with 14 other tissue types based on RNA-seq data. Darker red indicates higher relative reads per kilobase per million mapped reads (RPKM) counts. Mefs, mouse embryonic fibroblasts. (c) Relative expression (% of β-actin) of selected novel lncRNAs with associated ISEs in nine different tissues as determined by qPCR. Statistically significant differences were detected using a Kruskal–Wallis non-parametric test with a Dunn’s multiple comparison correction; *p<0.05, **p<0.01, ***p<0.001
Similar articles
- Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver.
Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, Bilenky M, Cezard T, Tam A, Kamoh B, Birol I, Dai D, Zhao Y, Hirst M, Verchere CB, Helgason CD, Marra MA, Jones SJ, Hoodless PA. Hoffman BG, et al. Genome Res. 2010 Aug;20(8):1037-51. doi: 10.1101/gr.104356.109. Epub 2010 Jun 15. Genome Res. 2010. PMID: 20551221 Free PMC article. - The study of regulatory effects of Pdx-1, MafA and NeuroD1 on the activity of porcine insulin promoter and the expression of human islet amyloid polypeptide.
Liu XD, Ruan JX, Xia JH, Yang SL, Fan JH, Li K. Liu XD, et al. Mol Cell Biochem. 2014 Sep;394(1-2):59-66. doi: 10.1007/s11010-014-2081-8. Epub 2014 May 14. Mol Cell Biochem. 2014. PMID: 24825179 - LIM domain-binding 1 maintains the terminally differentiated state of pancreatic β cells.
Ediger BN, Lim HW, Juliana C, Groff DN, Williams LT, Dominguez G, Liu JH, Taylor BL, Walp ER, Kameswaran V, Yang J, Liu C, Hunter CS, Kaestner KH, Naji A, Li C, Sander M, Stein R, Sussel L, Won KJ, May CL, Stoffers DA. Ediger BN, et al. J Clin Invest. 2017 Jan 3;127(1):215-229. doi: 10.1172/JCI88016. Epub 2016 Dec 12. J Clin Invest. 2017. PMID: 27941246 Free PMC article. - PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration.
Zhu Y, Liu Q, Zhou Z, Ikeda Y. Zhu Y, et al. Stem Cell Res Ther. 2017 Nov 2;8(1):240. doi: 10.1186/s13287-017-0694-z. Stem Cell Res Ther. 2017. PMID: 29096722 Free PMC article. Review. - Emerging roles of GLIS3 in neonatal diabetes, type 1 and type 2 diabetes.
Wen X, Yang Y. Wen X, et al. J Mol Endocrinol. 2017 Feb;58(2):R73-R85. doi: 10.1530/JME-16-0232. Epub 2016 Nov 29. J Mol Endocrinol. 2017. PMID: 27899417 Review.
Cited by
- Transcription factor GLIS3: Critical roles in thyroid hormone biosynthesis, hypothyroidism, pancreatic beta cells and diabetes.
Scoville DW, Kang HS, Jetten AM. Scoville DW, et al. Pharmacol Ther. 2020 Nov;215:107632. doi: 10.1016/j.pharmthera.2020.107632. Epub 2020 Jul 18. Pharmacol Ther. 2020. PMID: 32693112 Free PMC article. Review. - Role of transcription factors in the transdifferentiation of pancreatic islet cells.
van der Meulen T, Huising MO. van der Meulen T, et al. J Mol Endocrinol. 2015 Apr;54(2):R103-17. doi: 10.1530/JME-14-0290. J Mol Endocrinol. 2015. PMID: 25791577 Free PMC article. Review. - Coregulator Sin3a Promotes Postnatal Murine β-Cell Fitness by Regulating Genes in Ca2+ Homeostasis, Cell Survival, Vesicle Biosynthesis, Glucose Metabolism, and Stress Response.
Yang X, Graff SM, Heiser CN, Ho KH, Chen B, Simmons AJ, Southard-Smith AN, David G, Jacobson DA, Kaverina I, Wright CVE, Lau KS, Gu G. Yang X, et al. Diabetes. 2020 Jun;69(6):1219-1231. doi: 10.2337/db19-0721. Epub 2020 Apr 3. Diabetes. 2020. PMID: 32245798 Free PMC article. - Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic β-cell function.
Jia S, Ivanov A, Blasevic D, Müller T, Purfürst B, Sun W, Chen W, Poy MN, Rajewsky N, Birchmeier C. Jia S, et al. EMBO J. 2015 May 12;34(10):1417-33. doi: 10.15252/embj.201490819. Epub 2015 Mar 31. EMBO J. 2015. PMID: 25828096 Free PMC article. - Induction of α cell-restricted Gc in dedifferentiating β cells contributes to stress-induced β-cell dysfunction.
Kuo T, Damle M, González BJ, Egli D, Lazar MA, Accili D. Kuo T, et al. JCI Insight. 2019 May 23;5(13):e128351. doi: 10.1172/jci.insight.128351. JCI Insight. 2019. PMID: 31120862 Free PMC article.
References
- Zaret KS. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet. 2008;9:329–340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Z01 ES101765/ImNIH/Intramural NIH HHS/United States
- HG005692/HG/NHGRI NIH HHS/United States
- MOP-102628/CAPMC/ CIHR/Canada
- R01 HG005692/HG/NHGRI NIH HHS/United States
- RMF-111626/CAPMC/ CIHR/Canada
- Z01 ES101765-04/ImNIH/Intramural NIH HHS/United States
- MOP-111010/CAPMC/ CIHR/Canada
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials