Exaggerated translation causes synaptic and behavioural aberrations associated with autism - PubMed (original) (raw)
. 2013 Jan 17;493(7432):411-5.
doi: 10.1038/nature11782. Epub 2012 Dec 23.
Affiliations
- PMID: 23263185
- PMCID: PMC3548017
- DOI: 10.1038/nature11782
Exaggerated translation causes synaptic and behavioural aberrations associated with autism
Emanuela Santini et al. Nature. 2013.
Abstract
Autism spectrum disorders (ASDs) are an early onset, heterogeneous group of heritable neuropsychiatric disorders with symptoms that include deficits in social interaction skills, impaired communication abilities, and ritualistic-like repetitive behaviours. One of the hypotheses for a common molecular mechanism underlying ASDs is altered translational control resulting in exaggerated protein synthesis. Genetic variants in chromosome 4q, which contains the EIF4E locus, have been described in patients with autism. Importantly, a rare single nucleotide polymorphism has been identified in autism that is associated with increased promoter activity in the EIF4E gene. Here we show that genetically increasing the levels of eukaryotic translation initiation factor 4E (eIF4E) in mice results in exaggerated cap-dependent translation and aberrant behaviours reminiscent of autism, including repetitive and perseverative behaviours and social interaction deficits. Moreover, these autistic-like behaviours are accompanied by synaptic pathophysiology in the medial prefrontal cortex, striatum and hippocampus. The autistic-like behaviours displayed by the eIF4E-transgenic mice are corrected by intracerebroventricular infusions of the cap-dependent translation inhibitor 4EGI-1. Our findings demonstrate a causal relationship between exaggerated cap-dependent translation, synaptic dysfunction and aberrant behaviours associated with autism.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Figure 1. eIF4E transgenic mice exhibit increased eIF4E/eIF4G interactions and exaggerated cap-dependent translation
a, eIF4E transgenic mice (4E Tg) exhibit increased eIF4E expression in multiple brain regions. n=4 mice/genotype, *p<0.05 vs wild-type (WT), Student's _t_-test. b, 4E Tg mice exhibit normal expression of other translational control proteins. n=4 mice/genotype. *p<0.05 vs WT, Student's _t_-test. c, 4E Tg mice exhibit increased eIF4E/eIF4G interactions. Immunoprecipitation of eIF4E (left panel) and eIF4G (right panel). n=3 mice/genotype, *p<0.05 vs WT, Student's _t_-test. d, 4E Tg mice exhibit increased translation as measured with SUnSET (see Supplementary Methods). Vertical line traces of each autoradiogram are shown on the right. n=3 mice/genotype, *p<0.05 vs WT, Student's _t_-test. – represents a control sample without puromycin. All data are shown as mean ± SEM.
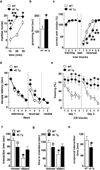
Figure 2. eIF4E transgenic mice exhibit ASD-like behaviors
eIF4E transgenic mice (4E Tg) were compared to wild-type (WT) littermates. a, Marble burying test: n=21–22 mice/genotype, ***p<0.001 and *p<0.05 vs WT, repeated measures (RM) ANOVA [time X genotype, F(2,46)=31.62, p< 0.001] followed by Bonferroni-Dunn test. b, Self-grooming test: n=12 mice/genotype, *p<0.05 vs WT, Student’s t_-test. c, Y-maze reversal task: n=21–22 mice/genotype, *p<0.05 and ***p<0.001 vs WT, RM ANOVA [time X genotype, F(5,138)=16.74, p< 0.001] followed by Bonferroni-Dunn test. d, Morris water maze (MWM) reversal learning: n=12–13 mice/genotype, *p<0.05 vs WT, RM ANOVA [time X genotype, F(3,92)=6.1, p< 0.001] followed by Bonferroni-Dunn test. e, Extinction of cued fear memory (15 CS/day represented as 3 CS/block): n=6 mice/genotype, *p<0.05 vs WT, RM ANOVA [day1: time X genotype, F(4,40)=5.73, p< 0.001; day2: time X genotype, F(4,40)=4.81, p< 0.01] followed by Bonferroni-Dunn test. f,g, Social behavior test: time spent interacting with either a stranger mouse f, or in the chambers g. n=6 mice/genotype, *p< 0.05 and °_p< 0.05 vs. WT, RM ANOVA [f: stimulus X genotype, F(1,10)=6.04, p< 0.05; g: stimulus X genotype, F(1,10)=6.12, p< 0.05] followed by Bonferroni-Dunn test. h, Reciprocal social interaction task: n=6 mice/genotype, *p<0.05 vs WT controls, Student's _t_-test. All data are shown as mean ± SEM.
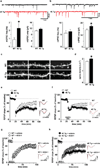
Figure 3. eIF4E transgenic mice exhibit alterations in synaptic function, dendritic spine density, and synaptic plasticity
a, eIF4E transgenic mice (4E Tg) exhibit increased mEPSC frequency and b, increased mIPSC amplitude in layer 2/3 mPFC pyramidal neurons. n=27–30 neurons/genotype, *p<0.05 vs WT, Student's _t_-test. c,d, 4E Tg mice exhibit increased dendritic spine density in layer 2/3 mPFC pyramidal neurons. High-magnification images c, and quantification d, of spiny dendrites from wild-type (WT) and 4E Tg mice. n= 12 neurons/genotype, *p<0.05 vs WT, Student's _t_-test. Scale bar = 2 µm. e, 4E Tg mice exhibit enhanced striatal LTD. n=13 slices from 8 mice/genotype. f, 4E Tg mice exhibit enhanced hippocampal mGluR-LTD. n=15 slices from 8 mice/genotype. g, h, 4EGI-1 normalizes enhanced striatal LTD displayed by 4E Tg mice h, without impacting LTD in WT mice g. n=18 slices from 9 mice/genotype/treatment. All field recordings were analyzed with repeated measure ANOVAs. Arrows indicate delivery of high-frequency stimulation (HFS). Solid bars indicate the duration of bath application of DHPG (10 µM, 10 min) and 4EGI-1 (100 µM, 45 min). Representative traces (right panels) showing fEPSP before (black) and 60 min after (red) HFS. All data are shown as mean ± SEM.
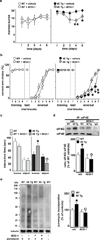
Figure 4. The cap-dependent translation inhibitor 4EGI-1 reverses ASD-like behaviors displayed by eIF4E transgenic mice
a, Treatment of eIF4E transgenic mice (4E Tg) with 4EGI-1 reduces the marble-burying behavior n=6 mice/genotype/treatment, **p< 0.01, *p< 0.05 vs. 4E Tg + vehicle, two-way repeated measures (RM) ANOVA [treatment X genotype, F(1,20)= 4.21, p< 0.05] followed by Bonferroni-Dunn test. b, 4EGI-1 improves the cognitive flexibility of 4E Tg mice in the Y-maze test. n=6–7 mice/genotype/treatment, **p< 0.01, *p< 0.05 vs. 4E Tg + vehicle, two-way RM ANOVA [treatment X genotype, F(1,21)=4.61, p< 0.05] followed by Bonferroni-Dunn test. c, 4EGI-1 improves social behavior of 4E Tg mice in the three-chamber arena test. n=6 mice/genotype/treatment, *p< 0.05 and °p< 0.05 vs. 4E Tg + vehicle, two-way RM ANOVA [treatment X genotype, F(1,20)=6.26, p< 0.05] followed by Bonferroni-Dunn test. d, 4EGI-1decreases the enhanced eIF4E/eIF4G interactions in 4E Tg mice. Immunoprecipitation (IP) of eIF4E in the striatum. n=4 mice/genotype, *p<0.05 and °p<0.05 vs vehicle-treated wild-type (WT) and 4E Tg, respectively, two-way ANOVA, followed by Bonferroni-Dunn test. e, 4EGI-1 normalizes the exaggerated cap-dependent translation in 4E Tg mice as measured with SUnSET. The last WT sample represents a control without puromycin. *p<0.05 and °p<0.05 vs vehicle-treated WT and 4E Tg, respectively, two-way ANOVA, followed by Bonferroni-Dunn test. All data are shown as mean ± SEM.
Comment in
- Neurodevelopmental disorders: Overproducing autism.
Yates D. Yates D. Nat Rev Neurosci. 2013 Feb;14(2):79. doi: 10.1038/nrn3440. Epub 2013 Jan 17. Nat Rev Neurosci. 2013. PMID: 23324663 No abstract available.
Similar articles
- Autism-related deficits via dysregulated eIF4E-dependent translational control.
Gkogkas CG, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, Vasuta C, Yee S, Truitt M, Dallaire P, Major F, Lasko P, Ruggero D, Nader K, Lacaille JC, Sonenberg N. Gkogkas CG, et al. Nature. 2013 Jan 17;493(7432):371-7. doi: 10.1038/nature11628. Epub 2012 Nov 21. Nature. 2013. PMID: 23172145 Free PMC article. - Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G.
Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, Halperin JA, Wagner G. Moerke NJ, et al. Cell. 2007 Jan 26;128(2):257-67. doi: 10.1016/j.cell.2006.11.046. Cell. 2007. PMID: 17254965 - Tuning Specific Translation in Cancer Metastasis and Synaptic Memory: Control at the MNK-eIF4E Axis.
Bramham CR, Jensen KB, Proud CG. Bramham CR, et al. Trends Biochem Sci. 2016 Oct;41(10):847-858. doi: 10.1016/j.tibs.2016.07.008. Epub 2016 Aug 12. Trends Biochem Sci. 2016. PMID: 27527252 Review. - Contrasting mechanisms of regulating translation of specific Drosophila germline mRNAs at the level of 5'-cap structure binding.
Lasko P, Cho P, Poulin F, Sonenberg N. Lasko P, et al. Biochem Soc Trans. 2005 Dec;33(Pt 6):1544-6. doi: 10.1042/BST0331544. Biochem Soc Trans. 2005. PMID: 16246166 Review.
Cited by
- Autism As a Disorder of High Intelligence.
Crespi BJ. Crespi BJ. Front Neurosci. 2016 Jun 30;10:300. doi: 10.3389/fnins.2016.00300. eCollection 2016. Front Neurosci. 2016. PMID: 27445671 Free PMC article. - Translating from cancer to the brain: regulation of protein synthesis by eIF4F.
Kats IR, Klann E. Kats IR, et al. Learn Mem. 2019 Aug 15;26(9):332-342. doi: 10.1101/lm.050047.119. Print 2019 Sep. Learn Mem. 2019. PMID: 31416906 Free PMC article. Review. - Neuroligin-3: A Circuit-Specific Synapse Organizer That Shapes Normal Function and Autism Spectrum Disorder-Associated Dysfunction.
Uchigashima M, Cheung A, Futai K. Uchigashima M, et al. Front Mol Neurosci. 2021 Oct 6;14:749164. doi: 10.3389/fnmol.2021.749164. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34690695 Free PMC article. Review. - Signalling pathways in autism spectrum disorder: mechanisms and therapeutic implications.
Jiang CC, Lin LS, Long S, Ke XY, Fukunaga K, Lu YM, Han F. Jiang CC, et al. Signal Transduct Target Ther. 2022 Jul 11;7(1):229. doi: 10.1038/s41392-022-01081-0. Signal Transduct Target Ther. 2022. PMID: 35817793 Free PMC article. Review. - Excitatory neuron-specific suppression of the integrated stress response contributes to autism-related phenotypes in fragile X syndrome.
Hooshmandi M, Sharma V, Thörn Perez C, Sood R, Krimbacher K, Wong C, Lister KC, Ureña Guzmán A, Bartley TD, Rocha C, Maussion G, Nadler E, Roque PM, Gantois I, Popic J, Lévesque M, Kaufman RJ, Avoli M, Sanz E, Nader K, Hagerman RJ, Durcan TM, Costa-Mattioli M, Prager-Khoutorsky M, Lacaille JC, Martinez-Cerdeno V, Gibson JR, Huber KM, Sonenberg N, Gkogkas CG, Khoutorsky A. Hooshmandi M, et al. Neuron. 2023 Oct 4;111(19):3028-3040.e6. doi: 10.1016/j.neuron.2023.06.017. Epub 2023 Jul 19. Neuron. 2023. PMID: 37473758 Free PMC article.
References
- Rapin I, Tuchman RF. Autism: definition, neurobiology, screening, diagnosis. Pediatric Clinics of NA. 2008;55:1129–1146. viii. - PubMed
- Kelleher RJ, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS078718/NS/NINDS NIH HHS/United States
- R01 CA154916/CA/NCI NIH HHS/United States
- R01 NS034007/NS/NINDS NIH HHS/United States
- CA154916/CA/NCI NIH HHS/United States
- R01 NS047384/NS/NINDS NIH HHS/United States
- NS047384/NS/NINDS NIH HHS/United States
- R37 NS034007/NS/NINDS NIH HHS/United States
- R29 NS034007/NS/NINDS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- NS034007/NS/NINDS NIH HHS/United States
- R21 NS078718/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous