Acute and fractionated irradiation differentially modulate glioma stem cell division kinetics - PubMed (original) (raw)
Acute and fractionated irradiation differentially modulate glioma stem cell division kinetics
Xuefeng Gao et al. Cancer Res. 2013.
Abstract
Glioblastoma multiforme (GBM) is one of the most aggressive human malignancies with a poor patient prognosis. Ionizing radiation either alone or adjuvant after surgery is part of standard treatment for GBM but remains primarily noncurative. The mechanisms underlying tumor radioresistance are manifold and, in part, accredited to a special subpopulation of tumorigenic cells. The so-called glioma stem cells (GSC) are bestowed with the exclusive ability to self-renew and repopulate the tumor and have been reported to be less sensitive to radiation-induced damage through preferential activation of DNA damage checkpoint responses and increased capacity for DNA damage repair. During each fraction of radiation, non-stem cancer cells (CC) die and GSCs become enriched and potentially increase in number, which may lead to accelerated repopulation. We propose a cellular Potts model that simulates the kinetics of GSCs and CCs in glioblastoma growth and radiation response. We parameterize and validate this model with experimental data of the U87-MG human glioblastoma cell line. Simulations are conducted to estimate GSC symmetric and asymmetric division rates and explore potential mechanisms for increased GSC fractions after irradiation. Simulations reveal that in addition to their higher radioresistance, a shift from asymmetric to symmetric division or a fast cycle of GSCs following fractionated radiation treatment is required to yield results that match experimental observations. We hypothesize a constitutive activation of stem cell division kinetics signaling pathways during fractionated treatment, which contributes to the frequently observed accelerated repopulation after therapeutic irradiation.
©2012 AACR.
Conflict of interest statement
No potential conflicts of interest are disclosed.
Figures
Figure 1
Schematic of cell division fate model. (A) Glioma stem cells (GSC, red circle) divide with rate k, either symmetrically with probability ps or asymmetrically with probability 1−ps, where one daughter cell is a non-stem cancer cell (CC, green hexagon) with proliferation potential ρmax. (B) Non-stem cancer cells (CC, green hexagon) grow with rate k, and produce two CCs with decremented proliferation potential _ρ_−1 if _ρ_>0, or die if proliferation potential is exhausted (_ρ_=0). (C) Flowchart of simulation process.
Figure 2
Model parameterization. (A) Tracking of migrating U87-MG cells in vitro and re-plotted with a common origin (left panel). Average migration distance (right panel; mean ± s.d., n=24). (B) Tracking of migrating generalized cells in silico and re-plotted with a common origin (left panel). Average displacement distance (mean ± s.d., n=30). (C) In vitro growth of a U87-MG cell population at t=4, 9 and 15 days. (D) In silico growth of a parameterized cell population at t=4, 9 and 15 days. Color-coded are GSC (red), proliferating CC (green) and quiescent CC (blue). (E) Population expansion via self-metastases at t=15, 75 and 120 days. Circles are added to visualize separate population clusters. Arrows indicate the locations of GSCs. Parameters used in the present simulation include _ps_=0.02, _ρmax_=10. Figures in (D) and (E) labeled with star mark (*) are identical.
Figure 3
Clonogenic survival fraction of U87-MG in vitro after single-doses of irradiation (mean + s.d., n=3) and unconstrained best-fit curve with _α_=0.3859 and _β_=0.01148. Survival curves plotted for estimated radioprotection factors of GSC (_λ_=0.1376) and quiescent CC/GSC (_ξ_=0.5).
Figure 4
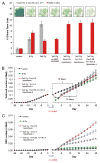
(A) Enrichment of CD133+ fraction in vitro (gray; means ± s.d., n=3) (22) and enrichment of GSC fraction in silico after irradiation for different GSC kinetics hypotheses (red; means ± s.d., n=5; see text for details). Representative in silico simulation snapshots are shown. Color-coded are GSC (red), proliferating CC (green) and quiescent CC (blue). Cell counts and simulation snapshots 48 hrs after irradiation. (B) Pre- and post-treatment total cell number evolution for different GSC kinetics hypotheses (means ± s.d., n=5). Treatment initiated at day 0. Curly brackets represent tumor population doubling times for control and irradiated tumors. (C) Pre- and post-treatment GSC number evolution for different GSC kinetics hypotheses (means ± s.d., n=5). Treatment initiated at day 0.
Figure 5
(A) Normal stem cell (SC) response to tissue injury adapted from (34). (B) Proposed GSC response to fractionated irradiation exposure.
References
- Glazier J, Graner F. Simulation of the differential adhesion driven rearrangement of biological cells. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1993;47(3):2128–54. - PubMed
- Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equation of State Calculations by Fast Computing Machines. J Chem Phys. 1953;21(6):1087.
- Cipra B. An introduction to the Ising model. The American Mathematical Monthly. 1987;94(10):937–59.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials