Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation - PubMed (original) (raw)
Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation
Asha Ivy Jacob et al. Mol Cell. 2013.
Abstract
Quality control of ribosomes is critical for cellular function since protein mistranslation leads to severe physiological consequences. We report evidence of a previously unrecognized ribosome quality control system in bacteria that operates at the level of 70S to remove defective ribosomes. YbeY, a previously unidentified endoribonuclease, and the exonuclease RNase R act together by a process mediated specifically by the 30S ribosomal subunit, to degrade defective 70S ribosomes but not properly matured 70S ribosomes or individual subunits. Furthermore, there is essentially no fully matured 16S rRNA in a ΔybeY mutant at 45°C, making YbeY the only endoribonuclease to be implicated in the critically important processing of the 16S rRNA 3' terminus. These key roles in ribosome quality control and maturation indicate why YbeY is a member of the minimal bacterial gene set and suggest that it could be a potential target for antibacterial drugs.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Figure 1. YbeY is a metal-dependent single strand specific endoribonuclease
Purified YbeY was used at a concentration of 5 μM in all assays unless mentioned otherwise. (A) YbeY is able to degrade total rRNA (3.8 μM) isolated from E. coli. tRNA (4.5 μM) is a relatively poor substrate for YbeY. When rRNA (3.8 μM) and tRNA (4.5 μM) were mixed, YbeY preferentially degraded the rRNA. YbeY is inhibited by 50 mM EDTA. RNase A (5 μM) was used as control to show degradation of the mixture of rRNA (3.8 μM) and tRNA (4.5 μM). The positions of the 23S, 16S, 5S rRNA and tRNA are indicated. (B) YbeY cleaves folA mRNA (4.0 μM) generated by in vitro transcription. Digestion products were analyzed by Synergel/agarose gel electrophoresis. (C)–(G) In vitro cleavage assay to identify the substrate requirement of YbeY using short synthetic oligoribonucleotides. Assays were performed using YbeY (5.0 μM) and 5'-32P-labeled oligoribonucleotides (5.0 μM); (C) ssRNA, (D) dsRNA, (E) dsRNA containing a single stranded extension at the 3', (F) hairpin substrate with perfectly base paired blunt ends, (G) ssRNAs 7, 10, 12, 20 and 30 nt long. Digestion products were analyzed by polyacrylamide gel electrophoresis. OH−, alkali ladder. ▼ indicates sites of cleavage by YbeY on the substrate (see also Figure S1 and S2).
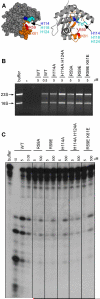
Figure 2. RNase activity of YbeY requires the histidine H114 residues and conserved amino acid residue R59
(A) Model of YbeY generated in PyMOL (
) using Swiss Prot Entry P77385 (Zhan et al., 2005), showing the positions of conserved residues R59, K61, H114, H118 and H124 in the proposed RNA channel (highlighted in orange). (B) – (C) RNase activities of YbeY wild type and mutant proteins (H114A; H114A H124A; R59A; R59E; R59E K61E) on (B) total rRNA (3.8 μM) isolated from E. coli and (C) a short 30 nt ssRNA substrate (5.0 μM) as shown in Figure 2A. Digestion products were analyzed by Synergel/agarose or polyacrylamide gel electrophoresis, respectively. RNase concentrations between 0.05 and 500 μM were used as indicated (see also Figure S3).
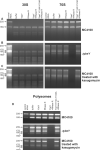
Figure 3. YbeY and RNase R together eliminate defective non-translating and translating ribosomes
Polysomes, 70S ribosomes and 30S and 50S ribosomal subunits were isolated from wild type MC4100 and Δ_ybeY_, and wild type MC4100 treated with 200 μg/ml kasugamycin. Ribosomal subunits (4.3 μM), 70S ribosomes (5.5 μM) and polysomes (3.9 μM) were incubated with equimolar amounts (5 μM each) of (i) YbeY, (ii) RNase R, (iii) YbeY and RNase R, and (iv) YbeY H114A H124A and RNase R as indicated. rRNA was thereafter extracted from these samples and analyzed on a Synergel/agarose gel. (A) rRNA from 30S ribosomal subunits and 70S ribosomes from MC4100 after incubation with YbeY and/or RNase R. Total rRNA isolated from wild type cells was used as a marker. (B) rRNA from 30S ribosomal subunits and 70S ribosomes from the Δ_ybeY_ mutant strain after incubation with YbeY and/or RNase R. Total rRNA isolated from the Δ_ybeY_ mutant strain was used as a marker. (C) rRNA from 30S ribosomal subunits and 70S ribosomes from kasugamycin-treated MC4100 after incubation with YbeY and/or RNase R. Total rRNA isolated from kasugamycin-treated wild type cells was used as a marker. (D) rRNA from polysomes isolated from MC4100, the Δ_ybeY_ mutant strain and MC4100 treated with 200 μg/ml kasugamycin after incubation with YbeY and/or RNase R. Total RNA from wild type cells, the Δ_ybeY_ mutant strain and wild type treated with 200 μg/ml kasugamycin, respectively, was used as marker (see also Figure S4).
Figure 4. Ribosome quality control requires 70S ribosome complex formation and is mediated by defective 30S ribosomal subunit
70S ribosomes isolated from the Δ_ybeY_ mutant were dissociated by fractionation on a sucrose gradient run at low magnesium concentration. (A) 50S and 30S ribosomal subunits (4.3 μM each) were subjected separately to digestion with YbeY and RNase R and the rRNA was analyzed on a Synergel/agarose gel. rRNA isolated from the Δ_ybeY_ mutant 70S ribosome was used as a marker (see also Figure S4). (B) 50S and 30S ribosomal subunits isolated from MC4100 and the Δ_ybeY_ mutant were mixed together in different combinations as indicated at equimolar concentrations (3.8 μM each) and subjected to digestion with YbeY and RNase R. rRNA was analyzed as described above.
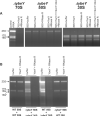
Figure 5. Thermosensitivity of the Δ_ybeY_ mutant can be attributed to the absence of mature 16S rRNA and strong defects in processing of the 16S rRNA 3' terminus
Cells were grown to an OD600 of 0.3 at 37°C and shifted to 45°C. Aliquots of cells were taken at time points indicated. (A) Total RNA extracted from MC4100 and the Δ_ybeY_ mutant grown at 37°C and 45°C at different time points after temperature shift was analyzed on a Synergel/agarose gel. Positions of the 23S, 17S, 16S and 16S* rRNA are indicated. (B) – (C) Ectopic expression of YbeY rescues the 16S rRNA phenotype in the Δ_ybeY_ mutant at 45°C. (B) Total RNA extracted from MC4100, MC4100 Δ_ybeY_ and MC4100 Δ_ybeY_ pYbeY grown at 37°C and 45°C was resolved on a Synergel/agarose gel. (C) The 5' and 3' termini of the 16S rRNA were mapped by primer extension and specific RNase H cleavage followed by Northern hybridization, respectively, as described (Davies et al., 2010). “P” and “ M” indicate the positions of bands derived from the precursor and mature forms of the rRNA (see also Figure S5).
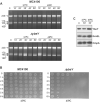
Figure 6. Severe 16S rRNA maturation defects of the Δ_ybeY_ mutant at 45°C are correlated with a substantial decrease in viability
(A) In the absence of new rRNA synthesis, almost no mature 16S rRNA is observed 20 min after the shift to 45°C in the Δ_ybeY_ mutant. MC4100 and the Δ_ybeY_ mutant were grown at 37°C to O.D600 of 0.3. Prior to temperature shift, rifampicin was added to the cultures at a concentration of 400 μg/ml to block transcription of new rRNA and cells collected at different time points after temperature shift from treated cells grown at 37°C and 45°C. Total RNA was extracted and analyzed on a Synergel/agarose gel. (B) – (C) Decrease in viability of the Δ_ybeY_ mutant within 1h exposure to 45°C and a two fold increase in YbeY levels under heat shock. (B) A detailed time course was performed on wild type E. coli MC4100 and the Δ_ybeY_ mutant at 45°C. Cells were grown to an OD600 of 0.3 at 37°C and shifted to 45°C. Aliquots of cells were taken at time points indicated, 10-fold serially diluted and 10 μl of each dilution were spotted on LB agar plates and incubated at 37°C. The Δ_ybeY_ mutant is severely compromised for growth at 45°C and there is a substantial decrease in viable cells within 1 hour of exposure to heat shock. (C) Levels of YbeY at 37°C and 45°C in MC4100 cells carrying a FLAG tagged genomic copy were determined by Western blot analysis using an anti-FLAG antibody. GroEL was used as control to ascertain the heat shock response and OmpA was used as a loading control (see also Figure S6).
Figure 7. Proposed model for ribosome quality control and 16S rRNA maturation by YbeY and RNase R. (A) Role of YbeY and RNase R in 70S ribosome quality control
YbeY acts as a sensor for defective 30S ribosomal subunits and, along with RNase R, exerts a key role in a unique quality control mechanism involving 70S ribosomes, but not individual 30S and 50S ribosomal subunits. After YbeY initiates the degradation of defective 70S ribosomes by making endonucleolytic nick/s in exposed single stranded portion/s of the rRNA, RNase R unwinds the RNA with its helicase activity (Awano et al., 2010) and continues the degradation of rRNA exonucleolytically, assisted further by YbeY. (B) Role of YbeY and RNase R in 16S rRNA maturation and/or rRNA stability: YbeY cleaves the 17S rRNA precursor endonucleolytically generating a 3' phosphate terminus at or near the final maturation site. Processing may be completed by the activity of additional RNases, such as RNase R, and/or a 3' phosphatase and may be further modulated by other ribosome maturation factors including Era, KsgA, RbfA or RsgA (Campbell and Brown, 2008; Inoue et al., 2006; Tu et al., 2009; Xu et al., 2008). Maturation of the 16S rRNA 3' terminus can stabilize and protect the rRNA making it inaccessible to further degradation by other housekeeping RNases.
Comment in
- YbeY: the jealous tailor.
Warner JR. Warner JR. Mol Cell. 2013 Feb 7;49(3):422-3. doi: 10.1016/j.molcel.2013.01.021. Mol Cell. 2013. PMID: 23395272
Similar articles
- Identification of YbeY-Protein Interactions Involved in 16S rRNA Maturation and Stress Regulation in Escherichia coli.
Vercruysse M, Köhrer C, Shen Y, Proulx S, Ghosal A, Davies BW, RajBhandary UL, Walker GC. Vercruysse M, et al. mBio. 2016 Nov 8;7(6):e01785-16. doi: 10.1128/mBio.01785-16. mBio. 2016. PMID: 27834201 Free PMC article. - Elevated Levels of Era GTPase Improve Growth, 16S rRNA Processing, and 70S Ribosome Assembly of Escherichia coli Lacking Highly Conserved Multifunctional YbeY Endoribonuclease.
Ghosal A, Babu VMP, Walker GC. Ghosal A, et al. J Bacteriol. 2018 Aug 10;200(17):e00278-18. doi: 10.1128/JB.00278-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29914987 Free PMC article. - The RNase YbeY Is Vital for Ribosome Maturation, Stress Resistance, and Virulence of the Natural Genetic Engineer Agrobacterium tumefaciens.
Möller P, Busch P, Sauerbrei B, Kraus A, Förstner KU, Wen TN, Overlöper A, Lai EM, Narberhaus F. Möller P, et al. J Bacteriol. 2019 May 8;201(11):e00730-18. doi: 10.1128/JB.00730-18. Print 2019 Jun 1. J Bacteriol. 2019. PMID: 30885931 Free PMC article. - Structural insights into cell cycle control by essential GTPase Era.
Ji X. Ji X. Postepy Biochem. 2016;62(3):335-342. Postepy Biochem. 2016. PMID: 28132488 Free PMC article. Review. - RNA folding and ribosome assembly.
Woodson SA. Woodson SA. Curr Opin Chem Biol. 2008 Dec;12(6):667-73. doi: 10.1016/j.cbpa.2008.09.024. Epub 2008 Oct 18. Curr Opin Chem Biol. 2008. PMID: 18935976 Free PMC article. Review.
Cited by
- The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis.
Liu J, Zhou W, Liu G, Yang C, Sun Y, Wu W, Cao S, Wang C, Hai G, Wang Z, Bock R, Huang J, Cheng Y. Liu J, et al. Plant Physiol. 2015 May;168(1):205-21. doi: 10.1104/pp.114.255000. Epub 2015 Mar 25. Plant Physiol. 2015. PMID: 25810095 Free PMC article. - Bacterial Longevity Requires Protein Synthesis and a Stringent Response.
Yin L, Ma H, Nakayasu ES, Payne SH, Morris DR, Harwood CS. Yin L, et al. mBio. 2019 Oct 15;10(5):e02189-19. doi: 10.1128/mBio.02189-19. mBio. 2019. PMID: 31615958 Free PMC article. - Methodologies for bacterial ribonuclease characterization using RNA-seq.
Broglia L, Le Rhun A, Charpentier E. Broglia L, et al. FEMS Microbiol Rev. 2023 Sep 5;47(5):fuad049. doi: 10.1093/femsre/fuad049. FEMS Microbiol Rev. 2023. PMID: 37656885 Free PMC article. Review. - A comparison of key aspects of gene regulation in Streptomyces coelicolor and Escherichia coli using nucleotide-resolution transcription maps produced in parallel by global and differential RNA sequencing.
Romero DA, Hasan AH, Lin YF, Kime L, Ruiz-Larrabeiti O, Urem M, Bucca G, Mamanova L, Laing EE, van Wezel GP, Smith CP, Kaberdin VR, McDowall KJ. Romero DA, et al. Mol Microbiol. 2014 Sep 30;94(5):963-87. doi: 10.1111/mmi.12810. Online ahead of print. Mol Microbiol. 2014. PMID: 25266672 Free PMC article. - Staphylococcal exoribonuclease YhaM destabilizes ribosomes by targeting the mRNA of a hibernation factor.
Lipońska A, Lee H, Yap MF. Lipońska A, et al. Nucleic Acids Res. 2024 Aug 27;52(15):8998-9013. doi: 10.1093/nar/gkae596. Nucleic Acids Res. 2024. PMID: 38979572 Free PMC article.
References
- Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS microbiology reviews. 2010;34:883–923. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM031030/GM/NIGMS NIH HHS/United States
- GM31030/GM/NIGMS NIH HHS/United States
- R56 GM017151/GM/NIGMS NIH HHS/United States
- GM17151/GM/NIGMS NIH HHS/United States
- F32 GM017151/GM/NIGMS NIH HHS/United States
- R37 GM017151/GM/NIGMS NIH HHS/United States
- P30 ES002109/ES/NIEHS NIH HHS/United States
- R01 GM017151/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases