Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes - PubMed (original) (raw)
. 2013 Apr;62(4):1227-37.
doi: 10.2337/db12-1474. Epub 2012 Dec 28.
Affiliations
- PMID: 23274896
- PMCID: PMC3609555
- DOI: 10.2337/db12-1474
Stimulation of autophagy improves endoplasmic reticulum stress-induced diabetes
Etty Bachar-Wikstrom et al. Diabetes. 2013 Apr.
Abstract
Accumulation of misfolded proinsulin in the β-cell leads to dysfunction induced by endoplasmic reticulum (ER) stress, with diabetes as a consequence. Autophagy helps cellular adaptation to stress via clearance of misfolded proteins and damaged organelles. We studied the effects of proinsulin misfolding on autophagy and the impact of stimulating autophagy on diabetes progression in Akita mice, which carry a mutation in proinsulin, leading to its severe misfolding. Treatment of female diabetic Akita mice with rapamycin improved diabetes, increased pancreatic insulin content, and prevented β-cell apoptosis. In vitro, autophagic flux was increased in Akita β-cells. Treatment with rapamycin further stimulated autophagy, evidenced by increased autophagosome formation and enhancement of autophagosome-lysosome fusion. This was associated with attenuation of cellular stress and apoptosis. The mammalian target of rapamycin (mTOR) kinase inhibitor Torin1 mimicked the rapamycin effects on autophagy and stress, indicating that the beneficial effects of rapamycin are indeed mediated via inhibition of mTOR. Finally, inhibition of autophagy exacerbated stress and abolished the anti-ER stress effects of rapamycin. In conclusion, rapamycin reduces ER stress induced by accumulation of misfolded proinsulin, thereby improving diabetes and preventing β-cell apoptosis. The beneficial effects of rapamycin in this context strictly depend on autophagy; therefore, stimulating autophagy may become a therapeutic approach for diabetes.
Figures
FIG. 1.
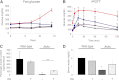
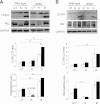
Rapamycin improves diabetes in Akita mice. Fed blood glucose (A), glucose tolerance (B), pancreatic insulin content (C), and serum insulin (D) of wild-type and Akita mice treated by daily IP injection of 0.2 g/kg rapamycin or the vehicle for 15 days. Animals underwent IP glucose tolerance test (IPGTT) by IP injection of 2 g/kg glucose at day 14. At termination, pancreatic insulin content and serum insulin were analyzed by RIA. Untreated wild-type mice, green; rapamycin-treated wild-type mice, purple; untreated Akita mice, red; and rapamycin-treated Akita mice, blue. Results are means ± SE of three mice in the wild-type groups and five in the Akita groups. *P < 0.05, **P < 0.01, #P < 0.001 for the difference between rapamycin-treated Akita mice and untreated controls (A and B) and between untreated wild-type and Akita mice (C and D).
FIG. 2.
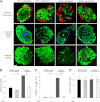
Rapamycin restores islet morphology, replenishes islet insulin content, and reduces β-cell turnover in diabetic Akita mice. Islet morphology (A), β-cell proliferation (A and B), apoptosis (A and C), and mass (D) in wild-type and Akita mice treated with and without rapamycin for 15 days. A: Pancreatic sections were immunostained for insulin, glucagon (top), and PDX1 (middle). Proliferation was assessed by staining for Ki67 (middle) and apoptosis by TUNEL assay (bottom). Scale bar, 20 μm. Total of 3,400–3,800 β-cells were counted per treatment (B_–_D). **P < 0.01, #P < 0.001 for the difference between the indicated groups. Rx, treatment with rapamycin.
FIG. 3.
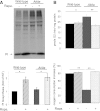
Rapamycin reduces cellular stress and prevents apoptosis in Akita β-cells. Akita and wild-type β-cells were treated with and without 50 nmol/L rapamycin (R) for 16 h, followed by analysis for different markers of insulin signaling and stress by Western blot and qPCR (spliced Xbp1 [_Xbp1s_]). Insulin signaling (IRS2, p-Akt/PKB, and p-S6) and UPR (A_–_C) and JNK and c-JUN phosphorylation (D and E). F: Apoptosis was assessed by Western blot for cleaved caspase 3 and quantification of cytosolic oligonucleosomes by ELISA (bottom). Representative blots and quantifications are shown. Results are means ± SE of three to five separate experiments. *P < 0.05, **P < 0.01, #P < 0.001 for the difference between the indicated groups. a.u., arbitrary units; Cl., cleaved; eIF2α, eukaryotic initiation factor 2α; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
FIG. 4.
Treatment with rapamycin increases proinsulin biosynthesis and glucose-stimulated oxygen consumption in Akita β-cells. Akita and wild-type β-cells were treated with and without 50 nmol/L rapamycin (Rapa.) for 16 h and then pulse-labeled for 45 min with [35_S_]-Met/Cys in KRBH-BSA buffer containing similar treatment. The labeling was terminated by ice-cold washout in glucose-free KRBH-BSA buffer. A: Proinsulin biosynthesis was determined by immunoprecipitation with anti-insulin serum (similar quantity of protein extracts were used for immunoprecipitation in the different experiments). Results are expressed as fold of proinsulin biosynthesis in untreated wild-type cells and are means ± SE of four individual experiments. B: Oxygen consumption was measured in wild-type and Akita cells in an XF24 respirometer. After basal measurements were recorded, cells were stimulated with 20 mmol/L glucose. Basal oxygen consumption is shown in the top panel and the increase in oxygen consumption in response to glucose in the bottom panel. *P < 0.05, **P < 0.01 for the difference between the indicated groups. PI, proinsulin.
FIG. 5.
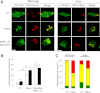
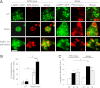
Rapamycin stimulates autophagy in wild-type and Akita β-cells. WT and Akita β-cells were cotransfected with LC3-GFP and P62-mCherry and then treated with and without rapamycin (Rapa.) for 16 h. Starved cells treated with the lysosome inhibitor bafilomycin A (Bafilo. A1) for 4 h were used as positive controls. Autophagosomes appear as LC3-GFP+ and/or P62-Cherry+ puncta (A). Quantification of number of autophagosomes per cell is shown in B (n = 10–13 cells per each treatment). Wild-type cells, white bars; Akita cells, black bars. C: Distribution of autophagosome types according to the expression of LC3-ΙΙ and P62/SQSTM1. Autophagosomes expressing LC3-ΙΙ or P62/SQSTM1 alone are shown in green and red, respectively; autophagosomes coexpressing LC3-ΙΙ and P62/SQSTM1 are shown in yellow (n = 10–13 cells/treatment). *P < 0.05, **P < 0.01, #P < 0.001 for the differences between the indicated groups (B) or between untreated (UT) Akita and wild-type cells or between rapamycin-treated cells and their matched controls (C). Scale bars, 5 μm.
FIG. 6.
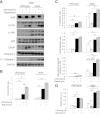
Western blot analysis of autophagy in Akita and wild-type β-cells (A) and islets (B). Wild-type and Akita β-cells and islets were treated without (UT) and with rapamycin (R) for 16 h or with bafilomycin A1 (B) for 4 h. Wild-type and Akita β-cells were incubated at 25 mmol/L glucose and islets at 22.2 mmol/L glucose. Representative blots of LC3-Ι/ΙΙ and P62/SQSTM1 expression and quantification of the experiments in wild-type and Akita β-cell lines are shown. Wild-type cells, white bars; Akita cells, black bars. Results are expressed as means ± SE of five separate experiments. *P < 0.05, **P < 0.01, #P < 0.001 for the difference between the indicated groups or between rapamycin-treated β-cells and their matched controls. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
FIG. 7.
Rapamycin (Rapa.) augments the generation of autolysosomes in wild-type and Akita β-cells. Autolysosome content was assessed using live-cell imaging of P62-mCherry and LAMP-1–GFP reporters. Wild-type and Akita β-cells were cotransfected with LAMP-1–GFP and P62-mCherry and then treated without (UT) and with rapamycin (R) for 16 h. Starved cells treated with the lysosome inhibitor bafilomycin A (B) were used as positive controls. Autolysosomes appear as P62-mCherry+/LAMP-1–GFP+ puncta (A). Quantification of autolysosomes, expressed as percentage of all P62-mCherry+ puncta, is shown (B) (n = 10–13 cells per each treatment). Wild-type cells, white bars; Akita cells, black bars. C: Quantification of autolysosome size (n = 10–13 cells per each treatment, at least 225 autolysosomes analyzed per treatment). *P < 0.05, **P < 0.01, #P < 0.001 for the difference between the indicated groups. Scale bar, 5 μm.
FIG. 8.
Inhibition of autophagy induces stress and apoptosis in β-cells and abolishes the beneficial effects of rapamycin in Akita β-cells. Wild-type and Akita β-cells were treated without and with rapamycin (50 nmol/L) and the lysosomal enzyme inhibitor chloroquine (50 μmol/L) for 16 h, followed by analysis for different markers of ER stress and apoptosis by Western blot (A). S6 phosphorylation was used as a marker for mTORC1 inhibition by rapamycin. Accumulation of LC3-ΙΙ in response to treatment with chloroquine indicates inhibition of autophagic flux (B). JNK phosphorylation, CHOP expression, and cleaved caspase 3 were used to assess stress and apoptosis (C). Quantification of apoptosis was performed by ELISA for cytosolic oligonucleosomes (D). Representative blots and quantifications are shown. Results are means ± SE of three to eight individual experiments. *P < 0.05, **P < 0.01, #P < 0.001 for the difference between the indicated groups. Cl., cleaved; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Similar articles
- Proinsulin misfolding and endoplasmic reticulum stress during the development and progression of diabetes.
Sun J, Cui J, He Q, Chen Z, Arvan P, Liu M. Sun J, et al. Mol Aspects Med. 2015 Apr;42:105-18. doi: 10.1016/j.mam.2015.01.001. Epub 2015 Jan 8. Mol Aspects Med. 2015. PMID: 25579745 Free PMC article. Review. - Improvement of ER stress-induced diabetes by stimulating autophagy.
Bachar-Wikstrom E, Wikstrom JD, Kaiser N, Cerasi E, Leibowitz G. Bachar-Wikstrom E, et al. Autophagy. 2013 Apr;9(4):626-8. doi: 10.4161/auto.23642. Epub 2013 Feb 4. Autophagy. 2013. PMID: 23380813 Free PMC article. - Glucose amplifies fatty acid-induced endoplasmic reticulum stress in pancreatic beta-cells via activation of mTORC1.
Bachar E, Ariav Y, Ketzinel-Gilad M, Cerasi E, Kaiser N, Leibowitz G. Bachar E, et al. PLoS One. 2009;4(3):e4954. doi: 10.1371/journal.pone.0004954. Epub 2009 Mar 23. PLoS One. 2009. PMID: 19305497 Free PMC article. - Misfolded proinsulin in the endoplasmic reticulum during development of beta cell failure in diabetes.
Arunagiri A, Haataja L, Cunningham CN, Shrestha N, Tsai B, Qi L, Liu M, Arvan P. Arunagiri A, et al. Ann N Y Acad Sci. 2018 Apr;1418(1):5-19. doi: 10.1111/nyas.13531. Epub 2018 Jan 28. Ann N Y Acad Sci. 2018. PMID: 29377149 Free PMC article. Review. - Autophagy plays a protective role in endoplasmic reticulum stress-mediated pancreatic β cell death.
Bartolome A, Guillen C, Benito M. Bartolome A, et al. Autophagy. 2012 Dec;8(12):1757-68. doi: 10.4161/auto.21994. Epub 2012 Sep 5. Autophagy. 2012. PMID: 22951927 Free PMC article.
Cited by
- Inhibition of autophagic turnover in β-cells by fatty acids and glucose leads to apoptotic cell death.
Mir SU, George NM, Zahoor L, Harms R, Guinn Z, Sarvetnick NE. Mir SU, et al. J Biol Chem. 2015 Mar 6;290(10):6071-85. doi: 10.1074/jbc.M114.605345. Epub 2014 Dec 29. J Biol Chem. 2015. PMID: 25548282 Free PMC article. - Proinsulin misfolding and endoplasmic reticulum stress during the development and progression of diabetes.
Sun J, Cui J, He Q, Chen Z, Arvan P, Liu M. Sun J, et al. Mol Aspects Med. 2015 Apr;42:105-18. doi: 10.1016/j.mam.2015.01.001. Epub 2015 Jan 8. Mol Aspects Med. 2015. PMID: 25579745 Free PMC article. Review. - A Selective Look at Autophagy in Pancreatic β-Cells.
Pearson GL, Gingerich MA, Walker EM, Biden TJ, Soleimanpour SA. Pearson GL, et al. Diabetes. 2021 Jun;70(6):1229-1241. doi: 10.2337/dbi20-0014. Epub 2021 May 20. Diabetes. 2021. PMID: 34016598 Free PMC article. Review. - Endoplasmic Reticulum Protein Quality Control in β Cells.
Shrestha N, Reinert RB, Qi L. Shrestha N, et al. Semin Cell Dev Biol. 2020 Jul;103:59-67. doi: 10.1016/j.semcdb.2020.04.006. Epub 2020 May 8. Semin Cell Dev Biol. 2020. PMID: 32402517 Free PMC article. Review. - INS-gene mutations: from genetics and beta cell biology to clinical disease.
Liu M, Sun J, Cui J, Chen W, Guo H, Barbetti F, Arvan P. Liu M, et al. Mol Aspects Med. 2015 Apr;42:3-18. doi: 10.1016/j.mam.2014.12.001. Epub 2014 Dec 24. Mol Aspects Med. 2015. PMID: 25542748 Free PMC article. Review.
References
- Van Lommel L, Janssens K, Quintens R, et al. Probe-independent and direct quantification of insulin mRNA and growth hormone mRNA in enriched cell preparations. Diabetes 2006;55:3214–3220 - PubMed
- Donath MY, Ehses JA, Maedler K, et al. Mechanisms of beta-cell death in type 2 diabetes. Diabetes 2005;54(Suppl. 2):S108–S113 - PubMed
- Eizirik DL, Cardozo AK, Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev 2008;29:42–61 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous