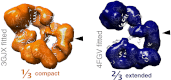Structural basis for cooperativity of CRM1 export complex formation - PubMed (original) (raw)
. 2013 Jan 15;110(3):960-5.
doi: 10.1073/pnas.1215214110. Epub 2012 Dec 31.
David Haselbach, Béla Voß, Andreas Russek, Piotr Neumann, Emma Thomson, Ed Hurt, Ulrich Zachariae, Holger Stark, Helmut Grubmüller, Achim Dickmanns, Ralf Ficner
Affiliations
- PMID: 23277578
- PMCID: PMC3549083
- DOI: 10.1073/pnas.1215214110
Structural basis for cooperativity of CRM1 export complex formation
Thomas Monecke et al. Proc Natl Acad Sci U S A. 2013.
Abstract
In eukaryotes, the nucleocytoplasmic transport of macromolecules is mainly mediated by soluble nuclear transport receptors of the karyopherin-β superfamily termed importins and exportins. The highly versatile exportin chromosome region maintenance 1 (CRM1) is essential for nuclear depletion of numerous structurally and functionally unrelated protein and ribonucleoprotein cargoes. CRM1 has been shown to adopt a toroidal structure in several functional transport complexes and was thought to maintain this conformation throughout the entire nucleocytoplasmic transport cycle. We solved crystal structures of free CRM1 from the thermophilic eukaryote Chaetomium thermophilum. Surprisingly, unbound CRM1 exhibits an overall extended and pitched superhelical conformation. The two regulatory regions, namely the acidic loop and the C-terminal α-helix, are dramatically repositioned in free CRM1 in comparison with the ternary CRM1-Ran-Snurportin1 export complex. Single-particle EM analysis demonstrates that, in a noncrystalline environment, free CRM1 exists in equilibrium between extended, superhelical and compact, ring-like conformations. Molecular dynamics simulations show that the C-terminal helix plays an important role in regulating the transition from an extended to a compact conformation and reveal how the binding site for nuclear export signals of cargoes is modulated by different CRM1 conformations. Combining these results, we propose a model for the cooperativity of CRM1 export complex assembly involving the long-range allosteric communication between the distant binding sites of GTP-bound Ran and cargo.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
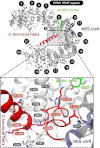
Crystal structure of free _ct_CRM1 (gray). The acidic loop (green), the C-terminal helix (red) and the NES cleft (blue) are highlighted. The HEAT repeats are numbered, and termini are labeled. (Lower) Rotated detail view of the interactions of residues from the C-terminal helix with the acidic loop and a patch of CRM1 formed by helices of H8 to H12. Hydrogen bonds and salt bridges are represented by dashed lines, and interacting residues are labeled.
Fig. 2.
Comparison of CRM1 conformations in different crystal structures. Free _ct_CRM1 (A), CRM1–RanGTP–SPN1 (B), CRM1–SPN1 (C), and CRM1–RanGTP–RanBP1 (D) are shown. CRM1 is depicted as rainbow colored surface from N (blue) to C terminus (red), whereas the interacting proteins are shown as tube models (SPN1, purple; RanGTP, beige; RanBP1, red). The position of the NES cleft in structures lacking cargo is marked by a black arrowhead. The bars at the side of the individual structures indicate the dimension of the protein in the shown orientation.
Fig. 3.
Comparison of NES cleft conformations between free _ct_CRM1 (PDB ID code 4FGV; green) and CRM1 bound to SPN1 and RanGTP (PDB ID code 3GJX; red). The NES cleft is shown in cartoon mode (Left) with the centers of mass of the helices 11A and 12A represented by blue spheres and their distances indicated. A surface model (Center) illustrates the differences of the NES clefts between both structures. A superposition of the SPN1 NES from the ternary CRM1–RanGTP–SPN1 complex (Right) highlights the structural changes in the NES cleft, which are incompatible with NES binding in free CRM1.
Fig. 4.
Single-particle EM analysis of free _ct_CRM1. EM models of the compact (orange) as well as the extended conformation (blue) of free _ct_CRM1 are shown. The crystal structures of free _ct_CRM1 and CRM1 in complex with SPN1 and RanGTP are fitted to the envelope models of the EM structures. The position of the NES cleft is marked by a black arrowhead.
Fig. 5.
MD simulations of WT and mutant free _ct_CRM1. Projections of WT simulations (A, cyan) and simulations with deleted C-terminal helix (B, red) onto the difference vector between the extended and compact structure constitute a measure of how much the protein changes into the compact conformation. (C) Projections onto the plane in the configurational space spanned by the extended, compact, and almost compact crystal structure show that, after deletion of the C-terminal helix, the system adopts the configuration of the almost compact structure (magenta square) rather than the compact conformation (orange square).
Fig. 6.
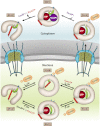
Model for cooperative CRM1 export complex assembly and disassembly showing its conformational variability and the important structural features in different states of the transport cycle. CRM1 is shown in the respective conformations and colored in gray with the acidic loop highlighted in green. The C-terminal helix of CRM1 is shown in red, and the NES binding cleft is represented by blue ovals. The PDB ID codes of the individual crystal structures used are indicated.
Similar articles
- Allosteric control of the exportin CRM1 unraveled by crystal structure analysis.
Monecke T, Dickmanns A, Ficner R. Monecke T, et al. FEBS J. 2014 Sep;281(18):4179-94. doi: 10.1111/febs.12842. Epub 2014 Jun 6. FEBS J. 2014. PMID: 24823279 Free PMC article. Review. - Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals.
Fung HY, Fu SC, Chook YM. Fung HY, et al. Elife. 2017 Mar 10;6:e23961. doi: 10.7554/eLife.23961. Elife. 2017. PMID: 28282025 Free PMC article. - A 2.1-Å-resolution crystal structure of unliganded CRM1 reveals the mechanism of autoinhibition.
Saito N, Matsuura Y. Saito N, et al. J Mol Biol. 2013 Jan 23;425(2):350-64. doi: 10.1016/j.jmb.2012.11.014. Epub 2012 Nov 16. J Mol Biol. 2013. PMID: 23164569 - Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex.
Petosa C, Schoehn G, Askjaer P, Bauer U, Moulin M, Steuerwald U, Soler-López M, Baudin F, Mattaj IW, Müller CW. Petosa C, et al. Mol Cell. 2004 Dec 3;16(5):761-75. doi: 10.1016/j.molcel.2004.11.018. Mol Cell. 2004. PMID: 15574331 - Atomic basis of CRM1-cargo recognition, release and inhibition.
Fung HY, Chook YM. Fung HY, et al. Semin Cancer Biol. 2014 Aug;27:52-61. doi: 10.1016/j.semcancer.2014.03.002. Epub 2014 Mar 12. Semin Cancer Biol. 2014. PMID: 24631835 Free PMC article. Review.
Cited by
- An integrated approach for genome annotation of the eukaryotic thermophile Chaetomium thermophilum.
Bock T, Chen WH, Ori A, Malik N, Silva-Martin N, Huerta-Cepas J, Powell ST, Kastritis PL, Smyshlyaev G, Vonkova I, Kirkpatrick J, Doerks T, Nesme L, Baßler J, Kos M, Hurt E, Carlomagno T, Gavin AC, Barabas O, Müller CW, van Noort V, Beck M, Bork P. Bock T, et al. Nucleic Acids Res. 2014 Dec 16;42(22):13525-33. doi: 10.1093/nar/gku1147. Epub 2014 Nov 14. Nucleic Acids Res. 2014. PMID: 25398899 Free PMC article. - Small GTP-binding protein Ran is regulated by posttranslational lysine acetylation.
de Boor S, Knyphausen P, Kuhlmann N, Wroblowski S, Brenig J, Scislowski L, Baldus L, Nolte H, Krüger M, Lammers M. de Boor S, et al. Proc Natl Acad Sci U S A. 2015 Jul 14;112(28):E3679-88. doi: 10.1073/pnas.1505995112. Epub 2015 Jun 29. Proc Natl Acad Sci U S A. 2015. PMID: 26124124 Free PMC article. - Karyopherin-mediated nucleocytoplasmic transport.
Wing CE, Fung HYJ, Chook YM. Wing CE, et al. Nat Rev Mol Cell Biol. 2022 May;23(5):307-328. doi: 10.1038/s41580-021-00446-7. Epub 2022 Jan 20. Nat Rev Mol Cell Biol. 2022. PMID: 35058649 Free PMC article. Review. - RanGTPase links nucleo-cytoplasmic transport to the recruitment of cargoes into small extracellular vesicles.
Chavan S, Khuperkar D, Lonare A, Panigrahi S, Bellare J, Rapole S, Seshadri V, Joseph J. Chavan S, et al. Cell Mol Life Sci. 2022 Jul 2;79(7):392. doi: 10.1007/s00018-022-04422-y. Cell Mol Life Sci. 2022. PMID: 35779171 Free PMC article. - Biallelic variants in SNUPN cause a limb girdle muscular dystrophy with myofibrillar-like features.
Iruzubieta P, Damborenea A, Ioghen M, Bajew S, Fernandez-Torrón R, Töpf A, Herrero-Reiriz Á, Epure D, Vill K, Hernández-Laín A, Manterola M, Azkargorta M, Pikatza-Menoio O, Pérez-Fernandez L, García-Puga M, Gaina G, Bastian A, Streata I, Walter MC, Müller-Felber W, Thiele S, Moragón S, Bastida-Lertxundi N, López-Cortajarena A, Elortza F, Gereñu G, Alonso-Martin S, Straub V, de Sancho D, Teleanu R, López de Munain A, Blázquez L. Iruzubieta P, et al. Brain. 2024 Aug 1;147(8):2867-2883. doi: 10.1093/brain/awae046. Brain. 2024. PMID: 38366623 Free PMC article.
References
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. - PubMed
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. - PubMed
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. - PubMed
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90(6):1041–1050. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous