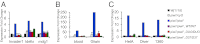Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity - PubMed (original) (raw)
Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity
Nicole Darricarrère et al. Proc Natl Acad Sci U S A. 2013.
Abstract
The Piwi protein subfamily is essential for Piwi-interacting RNA (piRNA) biogenesis, transposon silencing, and germ-line development, all of which have been proposed to require Piwi endonuclease activity, as validated for two cytoplasmic Piwi proteins in mice. However, recent evidence has led to questioning of the generality of this mechanism for the Piwi members that reside in the nucleus. Drosophila offers a distinct opportunity to study the function of nuclear Piwi proteins because, among three Drosophila Piwi proteins--called Piwi, Aubergine, and Argonaute 3--Piwi is the only member of this subfamily that is localized in the nucleus and expressed in both germ-line and somatic cells in the gonad, where it is responsible for piRNA biogenesis and regulatory functions essential for fertility. In this study, we demonstrate beyond doubt that the slicer activity of Piwi is not required for any known functions in vivo. We show that, in transgenic flies with the DDX catalytic triad of PIWI mutated, neither primary nor secondary piRNA biogenesis is detectably affected, transposons remain repressed, and fertility is normal. Our observations demonstrate that the mechanism of Piwi is independent of its in vitro endonuclease activity. Instead, it is consistent with the alternative mode of Piwi function as a molecule involved in the piRNA-directed guidance of epigenetic factors to chromatin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
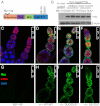
Fig. 1.
Slicer-deficient Piwi shows a normal level of expression and pattern of localization. (A) Schematic representation of the domain architecture of PIWI, highlighting the two critical aspartate residues essential for endonuclease activity, which were replaced by alanine in the slicer-deficient transgene. (B) Western blot showing similar levels of WT and DUO Piwi proteins in ovaries. The wild-type nontransgenic line (W 1118) is a negative control. Myc–Piwi transgenic line G38 was described (15). Protein levels of two independent site-specific transgenics are shown for wildtype Myc–Piwi at the attP2 site (WT) and Myc–Piwi slicer mutant (DUO). All transgenes are in the W 1118 ;piwi+ background. (C_–_J) Immunofluorescence micrographs of ovarioles from transgenic flies. Myc (green) was costained with Vasa (red) and DAPI (blue). C and G show no Myc staining in Myc-negative W 1118 ovaries. D and H show the normal localization of WT Myc–Piwi, whereas E and I show the normal localization of DUO Myc–PIWI (D614A, D685A). F and J show the localization of D37 Myc–PIWI (D614A, D685A).
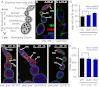
Fig. 2.
Slicer-deficient piwi mutants display normal germ-line stem cell division, oogenesis, and viability. (A) Schematic diagram of a normal fly ovariole. (B_–_F) Immunofluorescence images of ovarioles dissected from W[1118] strain (B), piwi 1 /piwi 1 mutant (C), piwi 1 /piwi 1 ;WT/WT (D), piwi 1 /piwi 1 ;DUO/DUO (E), and piwi 1 /piwi 1 ;D37/D37 (F). Vasa staining (red) highlights germ-line cells. 1B1 staining (green) outlines somatic cells, spectrosomes, and fusomes. DAPI reveals nuclear morphology. (G and H) Average fertility (G) and viability (H) of transgenic rescue fly strains are graphed with SE indicated. piwi 1 mutant flies fail to lay any egg due to ovarian defects as shown in C.
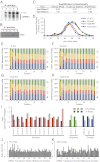
Fig. 3.
Duo mutant has no detectable defects in piRNA biogenesis. (A_–_C) Total levels of Piwi-associated piRNAs from +/+;WT/WT (lane 2), +/+;DUO/DUO (lane 3), piwi 1 /piwi 1;WT/WT (lane 4), and piwi 1 /piwi 1;DUO/DUO (lane 5) ovaries, resolved by denaturing PAGE (A). (B) Immunoprecipitation efficiency by Western blotting. (A and B) Lane 1 corresponds to W[1118], no-myc negative control. (C) Quantification by densitometry and normalization of piRNA/Piwi protein. (D) Size profile of ovarian piRNAs as revealed by deep sequencing. (E_–_H) Sequence profiles of piRNAs (∼23–28 nt) from IP libraries of piwi 1 /piwi 1 ;WT/WT (E) and piwi 1 /piwi 1 ;DUO/DUO (F) flies and from Total small RNA libraries of piwi 1 /piwi 1 ;WT/WT (G) and piwi 1 /piwi 1 ;DUO/DUO (H) flies. (I) Relative abundance of piRNAs derived from 17 known piRNA clusters from fly ovarian Piwi co-IP and total small RNA libraries, calculated as fold change of slicer mutant (DUO) over wildtype (WT). Only unique piRNAs are counted in this calculation, and the abundance is scaled to sequencing depth. The 17 clusters are grouped into three categories: germ-line–enriched (red), somatic-enriched (blue), and a ubiquitous class, comprising piRNAs similarly abundant in germ line and soma (green). Dark colors represent the Piwi co-IP libraries, and light colors represent the Total small RNA libraries. (J and K) Relative abundance of unique piRNAs derived from each transposon class in IP (J) and Total (K) small RNA libraries, shown as fold change of slicer mutant over wildtype. The abundance is scaled to the sequencing depth.
Fig. 4.
Piwi’s catalytic triad is not necessary to suppress transposons. The expression level of various transposons (invader1, idefix, mdg1, blood, gtwin, HetA, diver, and 1360) in ovaries was measured by qRT-PCR. After normalization of transposon levels per actin, the fold increase for each strain was calculated as a ratio over the wildtype W[1118] strain. The transposons are presented in three panels (A, B, and C) according to the magnitude of Piwi suppression towards their expression.
Similar articles
- DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary.
Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K. Ohtani H, et al. Genes Dev. 2013 Aug 1;27(15):1656-61. doi: 10.1101/gad.221515.113. Genes Dev. 2013. PMID: 23913921 Free PMC article. - Multiple roles for Piwi in silencing Drosophila transposons.
Rozhkov NV, Hammell M, Hannon GJ. Rozhkov NV, et al. Genes Dev. 2013 Feb 15;27(4):400-12. doi: 10.1101/gad.209767.112. Epub 2013 Feb 7. Genes Dev. 2013. PMID: 23392609 Free PMC article. - Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line.
Wang SH, Elgin SC. Wang SH, et al. Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21164-9. doi: 10.1073/pnas.1107892109. Epub 2011 Dec 12. Proc Natl Acad Sci U S A. 2011. PMID: 22160707 Free PMC article. - Beyond transposons: the epigenetic and somatic functions of the Piwi-piRNA mechanism.
Peng JC, Lin H. Peng JC, et al. Curr Opin Cell Biol. 2013 Apr;25(2):190-4. doi: 10.1016/j.ceb.2013.01.010. Epub 2013 Mar 4. Curr Opin Cell Biol. 2013. PMID: 23465540 Free PMC article. Review. - Untangling the web: the diverse functions of the PIWI/piRNA pathway.
Mani SR, Juliano CE. Mani SR, et al. Mol Reprod Dev. 2013 Aug;80(8):632-64. doi: 10.1002/mrd.22195. Epub 2013 Jun 27. Mol Reprod Dev. 2013. PMID: 23712694 Free PMC article. Review.
Cited by
- Maternally deposited germline piRNAs silence the tirant retrotransposon in somatic cells.
Akkouche A, Grentzinger T, Fablet M, Armenise C, Burlet N, Braman V, Chambeyron S, Vieira C. Akkouche A, et al. EMBO Rep. 2013 May;14(5):458-64. doi: 10.1038/embor.2013.38. Epub 2013 Apr 5. EMBO Rep. 2013. PMID: 23559065 Free PMC article. - Deficiency of MIWI2 (Piwil4) induces mouse erythroleukemia cell differentiation, but has no effect on hematopoiesis in vivo.
Jacobs JE, Wagner M, Dhahbi J, Boffelli D, Martin DI. Jacobs JE, et al. PLoS One. 2013 Dec 23;8(12):e82573. doi: 10.1371/journal.pone.0082573. eCollection 2013. PLoS One. 2013. PMID: 24376547 Free PMC article. - Zfrp8/PDCD2 is required in ovarian stem cells and interacts with the piRNA pathway machinery.
Minakhina S, Changela N, Steward R. Minakhina S, et al. Development. 2014 Jan;141(2):259-68. doi: 10.1242/dev.101410. Development. 2014. PMID: 24381196 Free PMC article. - From guide to target: molecular insights into eukaryotic RNA-interference machinery.
Ipsaro JJ, Joshua-Tor L. Ipsaro JJ, et al. Nat Struct Mol Biol. 2015 Jan;22(1):20-8. doi: 10.1038/nsmb.2931. Nat Struct Mol Biol. 2015. PMID: 25565029 Free PMC article. Review. - DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary.
Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K. Ohtani H, et al. Genes Dev. 2013 Aug 1;27(15):1656-61. doi: 10.1101/gad.221515.113. Genes Dev. 2013. PMID: 23913921 Free PMC article.
References
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. - PubMed
- Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases