Integrated analysis of gene expression profiles associated with response of platinum/paclitaxel-based treatment in epithelial ovarian cancer - PubMed (original) (raw)
Integrated analysis of gene expression profiles associated with response of platinum/paclitaxel-based treatment in epithelial ovarian cancer
Yong Han et al. PLoS One. 2012.
Abstract
Purpose: This study aims to explore gene expression signatures and serum biomarkers to predict intrinsic chemoresistance in epithelial ovarian cancer (EOC).
Patients and methods: Gene expression profiling data of 322 high-grade EOC cases between 2009 and 2010 in The Cancer Genome Atlas project (TCGA) were used to develop and validate gene expression signatures that could discriminate different responses to first-line platinum/paclitaxel-based treatments. A gene regulation network was then built to further identify hub genes responsible for differential gene expression between the complete response (CR) group and the progressive disease (PD) group. Further, to find more robust serum biomarkers for clinical application, we integrated our gene signatures and gene signatures reported previously to identify secretory protein-encoding genes by searching the DAVID database. In the end, gene-drug interaction network was constructed by searching Comparative Toxicogenomics Database (CTD) and literature.
Results: A 349-gene predictive model and an 18-gene model independent of key clinical features with high accuracy were developed for prediction of chemoresistance in EOC. Among them, ten important hub genes and six critical signaling pathways were identified to have important implications in chemotherapeutic response. Further, ten potential serum biomarkers were identified for predicting chemoresistance in EOC. Finally, we suggested some drugs for individualized treatment.
Conclusion: We have developed the predictive models and serum biomarkers for platinum/paclitaxel response and established the new approach to discover potential serum biomarkers from gene expression profiles. The potential drugs that target hub genes are also suggested.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
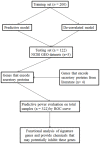
Figure 1. Work flow of the study design.
The 322 high-grade serous ovarian cancer cases were randomly divided in the training set (200 samples) and the testing set (122 samples). The training set was used to generate the predictive model and de-correlated model that is independent of key clinical features. Then these two models were validated using the testing set. Next we used 3 datasets from GEO database to validate signature genes in our findings. To explore potential biomarkers in serum, we combined signature genes in these two models with genes previously reported in four previous studies and queried these genes in DAVID database. Seventy-seven genes encoding secretory proteins were identified (Table S3). The predictability of those genes for chemotherapeutic response was then tested individually using the data from all 322 samples. Finally, we performed a functional analysis on those signature genes and suggested some drugs that could target the hub genes in our findings.
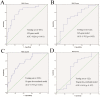
Figure 2. ROC curves of the two predictive models in the training set and the testing set.
(A) ROC curve of the 349-gene predictive model in training set (200 samples, AUC = 0.826; _p<_0.001. (B) ROC curve of the 349-gene predictive model in the testing set (122 samples, AUC = 0.702; p = 0.022). (C) ROC curve of the 18-gene de-correlated predictive model in the training set (200 samples, AUC = 0.775; p<0.001. (D) ROC curve of the 18-gene de-correlated predictive model in the testing set (122 samples, AUC = 0.614; p = 0.197).
Figure 3. Ten hub genes in the 349-gene signature.
Genes that interact with at least three other genes were selected, among which UBE2I, CASP3 and MAPK3 are important molecules that are involved in ovarian cancer progression or chemoresistance. Detailed information of these ten hub genes are listed in Table 4.
Figure 4. Hub genes and gene-gene interaction networks of top ten secretory protein-encoding genes.
(A) Hub genes and neighboring genes of top ten secretory protein-encoding genes. (B) AFM was exemplified to show potential mechanisms of the top ten secretory protein-encoding genes probably involving in chemoresistance.
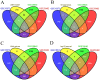
Figure 5. Venn diagram showing the overlap between our signatures genes and 3 external datasets from NCBI GEO database.
The Venn diagram shows how much genes in the 349-gene model (A), 18-gene model (B), hub genes (C), and top 10 serum biomarkers (D) are overlapped with 3 external datasets GSE15372, GSE28646 and GSE33482.
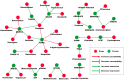
Figure 6. Hub gene-drug interaction network.
The hub gene-drug interaction network shows us how these genes and drugs could interact with each other. For example, ESR2 could increase the patient's susceptibility to Cisplatin, Etoposide and Raloxifene, while Gefitinib could increase the expression of ESR2.
Similar articles
- [Identification and prognostic value of differentially expressed proteins of patients with platinum resistance epithelial ovarian cancer in serum].
Wu WJ, Wang Q, Zhang W, Li L. Wu WJ, et al. Zhonghua Fu Chan Ke Za Zhi. 2016 Jul 25;51(7):515-23. doi: 10.3760/cma.j.issn.0529-567X.2016.07.007. Zhonghua Fu Chan Ke Za Zhi. 2016. PMID: 27465871 Chinese. - [Discrimination and clinical value of plasma metabolomic profiles in multidrug resistant epithelial ovarian cancer].
Wu WJ, Wang Q, Zhang W, Li L. Wu WJ, et al. Zhonghua Zhong Liu Za Zhi. 2017 Dec 23;39(12):896-902. doi: 10.3760/cma.j.issn.0253-3766.2017.12.004. Zhonghua Zhong Liu Za Zhi. 2017. PMID: 29262505 Chinese. - A combined blood based gene expression and plasma protein abundance signature for diagnosis of epithelial ovarian cancer--a study of the OVCAD consortium.
Pils D, Tong D, Hager G, Obermayr E, Aust S, Heinze G, Kohl M, Schuster E, Wolf A, Sehouli J, Braicu I, Vergote I, Van Gorp T, Mahner S, Concin N, Speiser P, Zeillinger R. Pils D, et al. BMC Cancer. 2013 Apr 3;13:178. doi: 10.1186/1471-2407-13-178. BMC Cancer. 2013. PMID: 23551967 Free PMC article. - [Docetaxel and ovarian cancer].
Ray-Coquard I. Ray-Coquard I. Bull Cancer. 2004 Feb;91(2):159-65. Bull Cancer. 2004. PMID: 15047455 Review. French. - Chemotherapy resistance in epithelial ovarian cancer: Mechanisms and emerging treatments.
Marchetti C, De Felice F, Romito A, Iacobelli V, Sassu CM, Corrado G, Ricci C, Scambia G, Fagotti A. Marchetti C, et al. Semin Cancer Biol. 2021 Dec;77:144-166. doi: 10.1016/j.semcancer.2021.08.011. Epub 2021 Aug 28. Semin Cancer Biol. 2021. PMID: 34464704 Review.
Cited by
- Eureka-DMA: an easy-to-operate graphical user interface for fast comprehensive investigation and analysis of DNA microarray data.
Abelson S. Abelson S. BMC Bioinformatics. 2014 Feb 24;15:53. doi: 10.1186/1471-2105-15-53. BMC Bioinformatics. 2014. PMID: 24564491 Free PMC article. - Identification of an Immune Gene-Based Cisplatin Response Model and CD27 as a Therapeutic Target against Cisplatin Resistance for Ovarian Cancer.
Guo Y, Yang N, Li G, Yin X, Dong L. Guo Y, et al. J Immunol Res. 2022 May 18;2022:4379216. doi: 10.1155/2022/4379216. eCollection 2022. J Immunol Res. 2022. PMID: 35647204 Free PMC article. - Identifying gene expression patterns associated with drug-specific survival in cancer patients.
Neary B, Zhou J, Qiu P. Neary B, et al. Sci Rep. 2021 Mar 2;11(1):5004. doi: 10.1038/s41598-021-84211-y. Sci Rep. 2021. PMID: 33654134 Free PMC article. - Exosome application in tumorigenesis: diagnosis and treatment of melanoma.
Karami Fath M, Azargoonjahromi A, Jafari N, Mehdi M, Alavi F, Daraei M, Mohammadkhani N, Mueller AL, Brockmueller A, Shakibaei M, Payandeh Z. Karami Fath M, et al. Med Oncol. 2022 Jan 4;39(2):19. doi: 10.1007/s12032-021-01621-8. Med Oncol. 2022. PMID: 34982284 Review. - Machine learning methods in the computational biology of cancer.
Vidyasagar M. Vidyasagar M. Proc Math Phys Eng Sci. 2014 Jul 8;470(2167):20140081. doi: 10.1098/rspa.2014.0081. Proc Math Phys Eng Sci. 2014. PMID: 25002826 Free PMC article. Review.
References
- Siegel R, Ward E, Brawley O, Jemal A (2011) Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 61: 212–236. - PubMed
- Vergote I, Trope CG, Amant F, Kristensen GB, Ehlen T, et al. (2010) Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N Engl J Med 363: 943–953. - PubMed
- McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, et al. (1996) Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med 334: 1–6. - PubMed
- Hartmann LC, Lu KH, Linette GP, Cliby WA, Kalli KR, et al. (2005) Gene expression profiles predict early relapse in ovarian cancer after platinum-paclitaxel chemotherapy. Clin Cancer Res 11: 2149–2155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
This study was funded by the National 973 Program of China (2009CB521805). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Medical