Regulation of miRNA biogenesis as an integrated component of growth factor signaling - PubMed (original) (raw)
Review
Regulation of miRNA biogenesis as an integrated component of growth factor signaling
Matthew T Blahna et al. Curr Opin Cell Biol. 2013 Apr.
Abstract
Transcriptional control of microRNAs (miRNA) by cell signaling pathways, especially in the context of growth factor regulation, is a widely recognized phenomenon with broad-reaching implications. However, several recent studies indicate that not just transcription, but also processing of miRNAs is subject to regulation as part of an integrated physiological response to various stimuli and environmental changes. The canonical miRNA biogenesis pathway; sequential steps of nucleolytic cleavage by the RNase III enzymes Drosha and Dicer, are emerging regulatory hubs for the modulation of miRNA expression as part of both physiological and pathological responses. In this article we use well-characterized growth-factor signaling pathways such as transforming growth factor-β (TGF-β), Protein Kinase B (PKB, also known as Akt) and extracellular-signal-regulated kinase (ERK) to illustrate how basic cell signaling pathways modulate the activities of these components of the miRNA biogenesis pathway to achieve optimal miRNA expression patterns.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
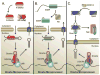
Figure 1. Basic components of the miRNA biogenesis pathway
miRNA begins as a long primary transcript (pri-miRNA) with a structure resembling mRNA. Pri-miRNAs contain a 5′ cap (m7-G) and a polyA tail at the 3′ end (AAAA). Pri-miRNA undergoes two sequential processing steps. First, the Drosha microprocessor complex which is composed of Drosha, DGCR8 and DEAD-Box RNA helicases (p68 or p72) cleaves pri-miRNA to generate precursor miRNA (pre-miRNA). Following cleavage by the Drosha microprocessor, the resulting pre-miRNA precursor maintains a stereotypical stem-loop structure. Following the export of pre-miRNA from the nucleus by exportin 5 (EXP5), Dicer processes the pre-miRNA into double stranded mature miRNA. Mature miRNA will be loaded into Argonaute proteins (Ago), which separates mature miRNA into two single stranded miRNAs. (B) Future miRNAs adopt a stereotypical stem-loop structure within the pri-miRNA sequence (shown here for miR-145). The mature sequence is encoded on one side of the loop and the degraded * strand on the other. More recent nomenclature has moved to calling each strand 5p and 3p according to which side of the strand they occupy. Here, the dotted lines indicate the site of future cleavage by Dicer and Drosha.
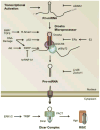
Figure 2. Integration of miRNA into cell growth factor signaling pathways
(A) Activation of receptor Smads by TGF-β family ligands leads to their translocation into the nucleus. In the nucleus, R-Smads bind, independent of the transcriptionally necessary co-Smad, to a conserved sequence in pri-miRNAs which they recruit to the Drosha microprocessor complex and facilitate pri- to pre-miRNA maturation. (B) PI3K-mediated activation of Akt induces the phosphorylation of KSRP. Phosphorylated KSRP is dissociated from mRNA and associates with targeted pri-miRNA sequence to recruit the Drosha microprocessor and facilitate pri- to pre-miRNA processing. (C) Activation of MAPK/ERK induces phophorylation of TRBP, which is subsequently recruited to Dicer and promotes the processing of miRNAs associated with cell proliferation.
Figure 3. Integration of biological signaling pathways at the sites of miRNA biogenesis
miRNA biogenesis pathways are emerging as critical components of gene regulatory mechanism controlled by growth factor signaling (and other) pathways. Activation or inactivation of the Drosha microprocessor by DNA binding proteins (Smads, p53, and ERα), RNA binding proteins (KSRP) or TBRP has been well documented. However, mechanisms of regulation of many other proteins in the miRNA biogenesis pathway by different signaling pathways remain to be uncovered.
Similar articles
- SMAD proteins control DROSHA-mediated microRNA maturation.
Davis BN, Hilyard AC, Lagna G, Hata A. Davis BN, et al. Nature. 2008 Jul 3;454(7200):56-61. doi: 10.1038/nature07086. Epub 2008 Jun 11. Nature. 2008. PMID: 18548003 Free PMC article. - Smad-mediated regulation of microRNA biosynthesis.
Blahna MT, Hata A. Blahna MT, et al. FEBS Lett. 2012 Jul 4;586(14):1906-12. doi: 10.1016/j.febslet.2012.01.041. Epub 2012 Jan 28. FEBS Lett. 2012. PMID: 22306316 Free PMC article. Review. - Smad-mediated miRNA processing: a critical role for a conserved RNA sequence.
Davis-Dusenbery BN, Hata A. Davis-Dusenbery BN, et al. RNA Biol. 2011 Jan-Feb;8(1):71-6. doi: 10.4161/rna.8.1.14299. Epub 2011 Jan 1. RNA Biol. 2011. PMID: 21289485 Free PMC article. Review. - Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis.
Kim YK, Kim B, Kim VN. Kim YK, et al. Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):E1881-9. doi: 10.1073/pnas.1602532113. Epub 2016 Mar 14. Proc Natl Acad Sci U S A. 2016. PMID: 26976605 Free PMC article. - miR-1306 Mediates the Feedback Regulation of the TGF-β/SMAD Signaling Pathway in Granulosa Cells.
Yang L, Du X, Liu L, Cao Q, Pan Z, Li Q. Yang L, et al. Cells. 2019 Mar 31;8(4):298. doi: 10.3390/cells8040298. Cells. 2019. PMID: 30935128 Free PMC article.
Cited by
- Effects of Pseudomonas aeruginosa on Microglial-Derived Extracellular Vesicle Biogenesis and Composition.
Jones LB, Kumar S, Bell CR, Peoples VA, Crenshaw BJ, Coats MT, Scoffield JA, Rowe GC, Sims B, Matthews QL. Jones LB, et al. Pathogens. 2019 Dec 14;8(4):297. doi: 10.3390/pathogens8040297. Pathogens. 2019. PMID: 31847332 Free PMC article. - Epstein-Barr Virus and miRNAs: Partners in Crime in the Pathogenesis of Multiple Sclerosis?
Hassani A, Khan G. Hassani A, et al. Front Immunol. 2019 Apr 3;10:695. doi: 10.3389/fimmu.2019.00695. eCollection 2019. Front Immunol. 2019. PMID: 31001286 Free PMC article. Review. - The regulation of microRNAs on chemoresistance in triple-negative breast cancer: a recent update.
Yan LJ, Y Lau AT, Xu YM. Yan LJ, et al. Epigenomics. 2024 Apr 19;16(8):571-87. doi: 10.2217/epi-2023-0430. Online ahead of print. Epigenomics. 2024. PMID: 38639712 Free PMC article. Review. - Isatin improves oligoasthenospermia caused by busulfan by regulating GSH/GPX4 axis to inhibit ferroptosis.
Wang C, Wang W, Dong J, Li X, Ye T, Zeng F, Jiang M, Shi J, Wang X, Zhang L. Wang C, et al. Front Pharmacol. 2024 Oct 31;15:1489956. doi: 10.3389/fphar.2024.1489956. eCollection 2024. Front Pharmacol. 2024. PMID: 39545065 Free PMC article. - Deregulation of Drosha in the pathogenesis of hereditary hemorrhagic telangiectasia.
Hata A, Lagna G. Hata A, et al. Curr Opin Hematol. 2019 May;26(3):161-169. doi: 10.1097/MOH.0000000000000493. Curr Opin Hematol. 2019. PMID: 30855334 Free PMC article. Review.
References
- Lee RC, Feinbaum RL, Ambros V. The C, elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
- van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nature reviews Cancer. 2011;11:644–656. - PubMed
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. - PubMed
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL093154/HL/NHLBI NIH HHS/United States
- R01 HL116191/HL/NHLBI NIH HHS/United States
- HL108317/HL/NHLBI NIH HHS/United States
- R01 HL108317/HL/NHLBI NIH HHS/United States
- HL093154/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous