Predicting functional effect of human missense mutations using PolyPhen-2 - PubMed (original) (raw)
Predicting functional effect of human missense mutations using PolyPhen-2
Ivan Adzhubei et al. Curr Protoc Hum Genet. 2013 Jan.
Abstract
PolyPhen-2 (Polymorphism Phenotyping v2), available as software and via a Web server, predicts the possible impact of amino acid substitutions on the stability and function of human proteins using structural and comparative evolutionary considerations. It performs functional annotation of single-nucleotide polymorphisms (SNPs), maps coding SNPs to gene transcripts, extracts protein sequence annotations and structural attributes, and builds conservation profiles. It then estimates the probability of the missense mutation being damaging based on a combination of all these properties. PolyPhen-2 features include a high-quality multiple protein sequence alignment pipeline and a prediction method employing machine-learning classification. The software also integrates the UCSC Genome Browser's human genome annotations and MultiZ multiple alignments of vertebrate genomes with the human genome. PolyPhen-2 is capable of analyzing large volumes of data produced by next-generation sequencing projects, thanks to built-in support for high-performance computing environments like Grid Engine and Platform LSF.
Figures
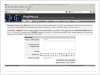
Figure 7.20.1
PolyPhen-2 home Web page with the input form prepared to submit a single protein substitution query using Swiss-Prot accession as a protein identifier. Also supported are RefSeq and Ensembl protein identifiers; alternatively, a dbSNP reference SNP identifier can be entered, in which case no other input is required.
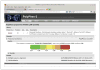
Figure 7.20.2
Detailed results of the PolyPhen-2 analysis for single variant query. This format is used for all PolyPhen-2 query reports except the Batch Query. The top Query section includes UniProtKB/Swiss-Prot description of query protein, if it was recognized as a known database entry. The large “heatmap” color bar with the black indicator mark dominates the display, illustrating the strength of the putative damaging effect for the variant, assessed using the default HumDiv-trained predictor. Clicking on the [+] control boxes expands the Prediction/Confidence panel for the HumDiv-trained predictor, as well as additional panels with protein multiple sequence alignment and 3D-structure viewers. For the color version of this figure, go to
http://www.currentprotocols.com/protocol/hg0720
.
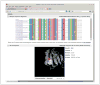
Figure 7.20.3
Detailed results of the PolyPhen-2 analysis for a single variant query with the multiple sequence alignment and 3-D-structure protein viewer panels expanded the multiple sequence alignment panel displays a fixed 75-residue wide window surrounding the variant’s position (the column indicated by black frame), with the alignment colored using the ClustalX (Thompson et al., 1997) scheme for all columns above 50% conservation threshold. Clicking on the link at the bottom of the alignment panel opens the Jalview (Waterhouse et al., 2009) alignment viewer applet with the complete multiple alignment loaded. Displayed below is a 3-D-structure viewer applet (Jmol;
) with the protein structure loaded and zoomed into the mutation residue using the Zoom into mutation button. The structure viewer window is fully interactive, and the protein structure can be rotated, moved, or zoomed in and out.
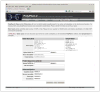
Figure 7.20.4
The PolyPhen-2 Batch Query Web page allows submitting large number of variants for analysis in a single operation. Type or paste your variants into the Batch Query text input area (one variant per line) or upload a text file listing variants using Upload batch file text box (locate the file using the Browse button). If you enter your e-mail address into the corresponding text box, you will be notified via e-mail when your query completes. To analyze protein variants in nonstandard or unannotated proteins, you can upload your own protein sequences in FASTA format using the Upload FASTA file text box. Genomic variants are also supported; see the Sample Batch panel for the various input format examples. Do not forget to select the genome assembly version matching your genomic SNP data under Advanced Options; default assembly version used is GRCh37/UCSC hg19.
Figure 7.20.5
Grid Gateway Interface (GGI) Web page showing a PolyPhen-2 user session with one single-variant query completed and a Batch Query pending execution. Click on the View link to access results of a single-variant query (no errors were reported). This Batch Query was queued as a 7-stage pipeline; the status of each pipeline stage is tracked and displayed separately, with short stage explanations printed in the Description column. The batch will be completed when the last stage finishes. Grid Status shows high Grid Load and large number of other Pending jobs; batch completion waiting time is likely to be substantial. Click on the Refresh button periodically to update session status. You can also close your browser and check your session at a later time—go to the PolyPhen-2 home page, click on the Check Status button, and you will be transferred to your session automatically.
Figure 7.20.6
Grid Gateway Interface (GGI) Web page showing a completed PolyPhen-2 Batch Query. Right-click on one of the SNPs, Short, or Full links in the Batches/Results column to download results to your computer; see Basic Protocol 2 for description of the three various types of report files. Click on the Logs link under Batches/Errors to view all error and warning messages generated by the pipeline. Note that most of the warnings are for your information only and do not indicate failure of the analysis. After downloading results, all batch data can be deleted by checking corresponding Batches/Delete checkbox and clicking on the Refresh button. Be warned that the delete operation is irreversible and deleted data cannot be restored.
Similar articles
- Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations.
Flanagan SE, Patch AM, Ellard S. Flanagan SE, et al. Genet Test Mol Biomarkers. 2010 Aug;14(4):533-7. doi: 10.1089/gtmb.2010.0036. Genet Test Mol Biomarkers. 2010. PMID: 20642364 - Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models.
Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR. Shihab HA, et al. Hum Mutat. 2013 Jan;34(1):57-65. doi: 10.1002/humu.22225. Epub 2012 Nov 2. Hum Mutat. 2013. PMID: 23033316 Free PMC article. - Machine learning classifier for identification of damaging missense mutations exclusive to human mitochondrial DNA-encoded polypeptides.
Martín-Navarro A, Gaudioso-Simón A, Álvarez-Jarreta J, Montoya J, Mayordomo E, Ruiz-Pesini E. Martín-Navarro A, et al. BMC Bioinformatics. 2017 Mar 7;18(1):158. doi: 10.1186/s12859-017-1562-7. BMC Bioinformatics. 2017. PMID: 28270093 Free PMC article. - Computational prediction of the effects of non-synonymous single nucleotide polymorphisms in human DNA repair genes.
Nakken S, Alseth I, Rognes T. Nakken S, et al. Neuroscience. 2007 Apr 14;145(4):1273-9. doi: 10.1016/j.neuroscience.2006.09.004. Epub 2006 Oct 19. Neuroscience. 2007. PMID: 17055652 Review. - Integrating Large-Scale Protein Structure Prediction into Human Genetics Research.
Correa Marrero M, Jänes J, Baptista D, Beltrao P. Correa Marrero M, et al. Annu Rev Genomics Hum Genet. 2024 Aug;25(1):123-140. doi: 10.1146/annurev-genom-120622-020615. Epub 2024 Aug 6. Annu Rev Genomics Hum Genet. 2024. PMID: 38621234 Review.
Cited by
- Informing patients about their mutation tests: CDKN2A c.256G>A in melanoma as an example.
Hemminki K, Srivastava A, Rachakonda S, Bandapalli O, Nagore E, Hemminki A, Kumar R. Hemminki K, et al. Hered Cancer Clin Pract. 2020 Jul 31;18:15. doi: 10.1186/s13053-020-00146-x. eCollection 2020. Hered Cancer Clin Pract. 2020. PMID: 32760473 Free PMC article. - Rare host variants in ciliary expressed genes contribute to COVID-19 severity in Bulgarian patients.
Kamenarova K, Kachakova-Yordanova D, Baymakova M, Georgiev M, Mihova K, Petkova V, Beltcheva O, Argirova R, Atanasov P, Kunchev M, Andonova R, Zasheva A, Drenska R, Ivanov I, Pantileeva D, Koleva V, Penev A, Lekova-Nikova D, Georgiev D, Pencheva D, Bozhilova R, Ivanova N, Dimova I, Plochev K, Popov G, Popivanov I, Gabrovsky N, Leseva M, Mitev V, Kaneva R. Kamenarova K, et al. Sci Rep. 2024 Aug 22;14(1):19487. doi: 10.1038/s41598-024-70514-3. Sci Rep. 2024. PMID: 39174791 Free PMC article. - Complete loss of the DNAJB6 G/F domain and novel missense mutations cause distal-onset DNAJB6 myopathy.
Ruggieri A, Brancati F, Zanotti S, Maggi L, Pasanisi MB, Saredi S, Terracciano C, Antozzi C, D Apice MR, Sangiuolo F, Novelli G, Marshall CR, Scherer SW, Morandi L, Federici L, Massa R, Mora M, Minassian BA. Ruggieri A, et al. Acta Neuropathol Commun. 2015 Jul 25;3:44. doi: 10.1186/s40478-015-0224-0. Acta Neuropathol Commun. 2015. PMID: 26205529 Free PMC article. - Phenotypic spectrum in recessive STING-associated vasculopathy with onset in infancy: Four novel cases and analysis of previously reported cases.
Wan R, Fänder J, Zakaraia I, Lee-Kirsch MA, Wolf C, Lucas N, Olfe LI, Hendrich C, Jonigk D, Holzinger D, Steindor M, Schmidt G, Davenport C, Klemann C, Schwerk N, Griese M, Schlegelberger B, Stehling F, Happle C, Auber B, Steinemann D, Wetzke M, von Hardenberg S. Wan R, et al. Front Immunol. 2022 Oct 6;13:1029423. doi: 10.3389/fimmu.2022.1029423. eCollection 2022. Front Immunol. 2022. PMID: 36275728 Free PMC article. - A Novel Missense Mutation in the Spectrin Beta Nonerythrocytic 2 Gene Likely Associated with Spinocerebellar Ataxia Type 5.
Liu LZ, Ren M, Li M, Ren YT, Sun B, Sun XS, Chen SY, Li SY, Huang XS. Liu LZ, et al. Chin Med J (Engl). 2016 Oct 20;129(20):2516-2517. doi: 10.4103/0366-6999.191834. Chin Med J (Engl). 2016. PMID: 27748352 Free PMC article. No abstract available.
References
- Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. - PMC - PubMed
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. - PubMed
- Chasman D, Adams RM. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: Structure-based assessment of amino acid variation. J Mol Biol. 2001;307:683–706. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources